7FFQ
 
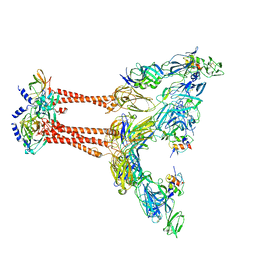 | | Cryo-EM structure of VEEV VLP at the 2-fold axes | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFE
 
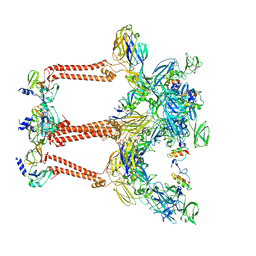 | | Cryo-EM structure of VEEV VLP | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFF
 
 | | Structure of Venezuelan equine encephalitis virus with the receptor LDLRAD3 | | Descriptor: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFN
 
 | | Cryo-EM structure of VEEV VLP-LDLRAD3-D1 complex at the 5-fold axes | | Descriptor: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFL
 
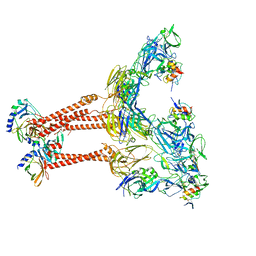 | | Cryo-EM structure of VEEV VLP-LDLRAD3-D1 complex at the 2-fold axes | | Descriptor: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
3KH8
 
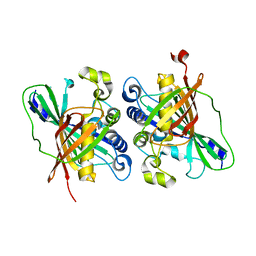 | | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici | | Descriptor: | MaoC-like dehydratase | | Authors: | Wang, H, Zhang, K, Guo, J, Zhou, Q, Zheng, X, Sun, F, Pang, H, Zhang, X. | | Deposit date: | 2009-10-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici
To be Published
|
|
5WWL
 
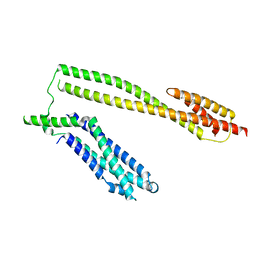 | | Crystal structure of the Schizogenesis pombe kinetochore Mis12C subcomplex | | Descriptor: | Centromere protein mis12, Kinetochore protein nnf1 | | Authors: | Wang, C, Zhou, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2017-01-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphorylation of CENP-C by Aurora B facilitates kinetochore attachment error correction in mitosis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTB
 
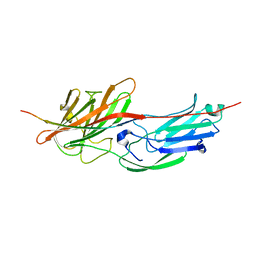 | | Complex Structure of Staphylococcus aureus SdrE with human complement factor H | | Descriptor: | Peptide from Complement factor H, Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
5J6P
 
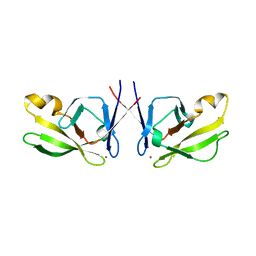 | | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe | | Descriptor: | Kinetochore protein mis18, ZINC ION | | Authors: | Wang, C, Shao, C, Zhang, M, Zhang, X, Zang, J. | | Deposit date: | 2016-04-05 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe
To Be Published
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
7M0R
 
 | | Cryo-EM structure of the Sema3A/PlexinA4/Neuropilin 1 complex | | Descriptor: | CALCIUM ION, Neuropilin-1, Plexin-A4, ... | | Authors: | Lu, D, Shang, G, He, X, Bai, X, Zhang, X. | | Deposit date: | 2021-03-11 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the Sema3A/PlexinA4/Neuropilin tripartite complex.
Nat Commun, 12, 2021
|
|
2R5W
 
 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase from Francisella tularensis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Li, X, Raffaelli, N, Grishin, N, Osterman, A, Zhang, H. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
2QJT
 
 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase complexed with AMP and MN ion from Francisella tularensis | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Raffaelli, N, Magni, G, Grishin, N.V, Osterman, A, Zhang, H. | | Deposit date: | 2007-07-09 | | Release date: | 2008-03-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
2QJO
 
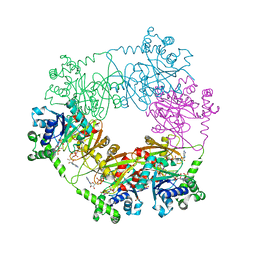 | | crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase (NadM) complexed with ADPRP and NAD from Synechocystis sp. | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Bifunctional NMN adenylyltransferase/Nudix hydrolase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Raffaelli, N, Magni, G, Grishin, N.V, Osterman, A, Zhang, H. | | Deposit date: | 2007-07-08 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
6LZ7
 
 | |
6LZ3
 
 | |
3ZN8
 
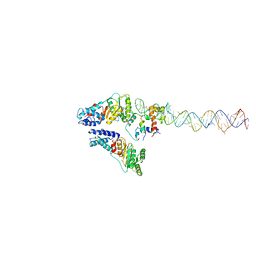 | | Structural Basis of Signal Sequence Surveillance and Selection by the SRP-SR Complex | | Descriptor: | 4.5 S RNA, DIPEPTIDYL AMINOPEPTIDASE B, MAGNESIUM ION, ... | | Authors: | von Loeffelholz, O, Knoops, K, Ariosa, A, Zhang, X, Karuppasamy, M, Huard, K, Schoehn, G, Berger, I, Shan, S.O, Schaffitzel, C. | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural Basis of Signal Sequence Surveillance and Selection by the Srp-Sr Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | Authors: | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
1Q0S
 
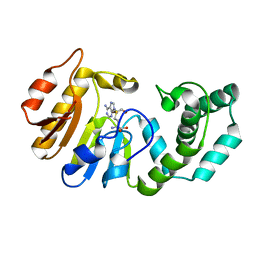 | | Binary Structure of T4DAM with AdoHcy | | Descriptor: | DNA adenine methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yang, Z, Horton, J.R, Zhou, L, Zhang, X.J, Dong, A, Zhang, X, Schlagman, S.L, Kossykh, V, Hattman, S, Cheng, X. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the bacteriophage T4 DNA adenine methyltransferase
Nat.Struct.Biol., 10, 2003
|
|
1Q0T
 
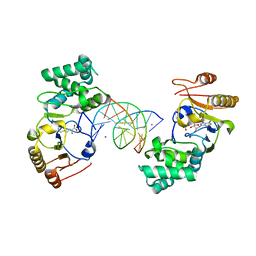 | | Ternary Structure of T4DAM with AdoHcy and DNA | | Descriptor: | 5'-D(*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*T)-3', DNA adenine methylase, IODIDE ION, ... | | Authors: | Yang, Z, Horton, J.R, Zhou, L, Zhang, X.J, Dong, A, Zhang, X, Schlagman, S.L, Kossykh, V, Hattman, S, Cheng, X. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the bacteriophage T4 DNA adenine methyltransferase
Nat.Struct.Biol., 10, 2003
|
|
1DDJ
 
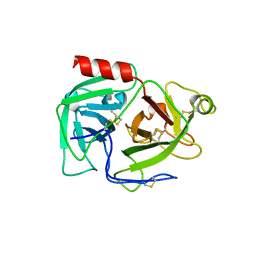 | | CRYSTAL STRUCTURE OF HUMAN PLASMINOGEN CATALYTIC DOMAIN | | Descriptor: | PLASMINOGEN | | Authors: | Wang, X, Terzyan, S, Tang, J, Loy, J, Lin, X, Zhang, X. | | Deposit date: | 1999-11-10 | | Release date: | 2000-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human plasminogen catalytic domain undergoes an unusual conformational change upon activation.
J.Mol.Biol., 295, 2000
|
|
4M8N
 
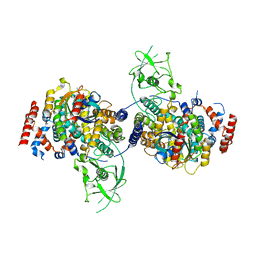 | | Crystal Structure of PlexinC1/Rap1B Complex | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pascoe, H.G, Wang, Y, Brautigam, C.A, He, H, Zhang, X. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin.
Elife, 2, 2013
|
|
7LLY
 
 | | Oxyntomodulin-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in complex with Gs protein | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wootten, D, Sexton, P.M, Belousoff, M.J, Danev, R, Zhang, X, Khoshouei, M, Venugopal, H. | | Deposit date: | 2021-02-04 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dynamics of GLP-1R peptide agonist engagement are correlated with kinetics of G protein activation.
Nat Commun, 13, 2022
|
|
7LLL
 
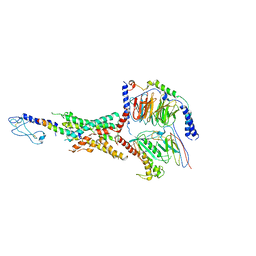 | | Exendin-4-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in complex with Gs protein | | Descriptor: | Exendin-4, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wootten, D, Sexton, P.M, Belousoff, M.J, Danev, R, Zhang, X, Khoshouei, M, Venugopal, H. | | Deposit date: | 2021-02-04 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Dynamics of GLP-1R peptide agonist engagement are correlated with kinetics of G protein activation.
Nat Commun, 13, 2022
|
|
4N26
 
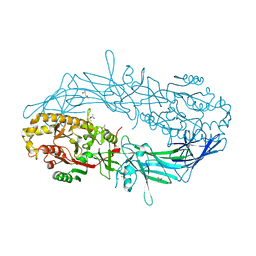 | | Crystal structure of Protein Arginine Deiminase 2 (500 uM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
