7WK7
 
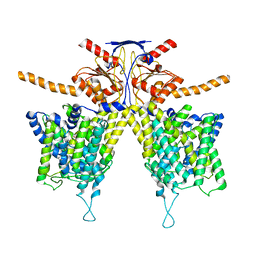 | | Mouse Pendrin bound bicarbonate in inward state | | Descriptor: | BICARBONATE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-08 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
6IYA
 
 | |
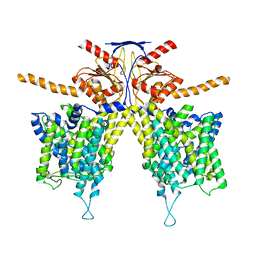
7WL8
 
 | |
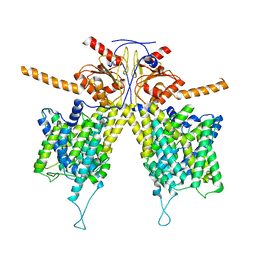
7WLB
 
 | |
8WTM
 
 | |
8WTP
 
 | |
8WTN
 
 | |
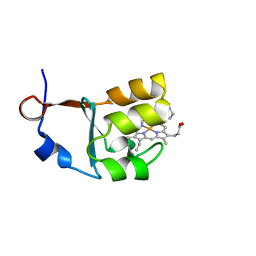
3OZZ
 
 | |
173L
 
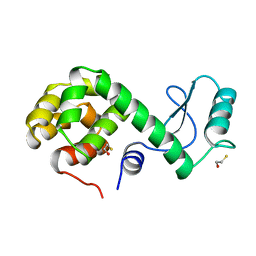 | | PROTEIN FLEXIBILITY AND ADAPTABILITY SEEN IN 25 CRYSTAL FORMS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Xiong, X.-P, Zhang, X.-J, Sun, D, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
3P16
 
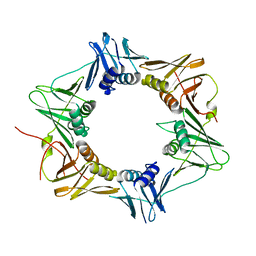 | | Crystal structure of DNA polymerase III sliding clamp | | Descriptor: | DNA polymerase III subunit beta | | Authors: | Gui, W.J, Lin, S.Q, Chen, Y.Y, Zhang, X.E, Bi, L.J, Jiang, T. | | Deposit date: | 2010-09-30 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of DNA polymerase III beta sliding clamp from Mycobacterium tuberculosis.
Biochem.Biophys.Res.Commun., 405, 2011
|
|
2IBJ
 
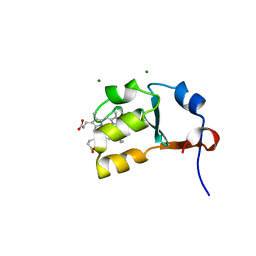 | | Structure of House Fly Cytochrome B5 | | Descriptor: | Cytochrome b5, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Terzyan, S, Zhang, X.C, Benson, D.R, Wang, L. | | Deposit date: | 2006-09-11 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparison of cytochromes b(5) from insects and vertebrates.
Proteins, 67, 2007
|
|
2DI4
 
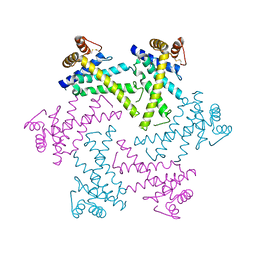 | | Crystal structure of the FtsH protease domain | | Descriptor: | Cell division protein ftsH homolog, MERCURY (II) ION | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-28 | | Release date: | 2006-06-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
8I38
 
 | |
8I3A
 
 | |
2J48
 
 | |
6VXK
 
 | | Cryo-EM Structure of the full-length A39R/PlexinC1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-C1, Semaphorin-like protein 139 | | Authors: | Kuo, Y.-C, Chen, H, Shang, G, Uchikawa, E, Tian, H, Bai, X, Zhang, X. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the PlexinC1/A39R complex reveals inter-domain interactions critical for ligand-induced activation.
Nat Commun, 11, 2020
|
|
2I89
 
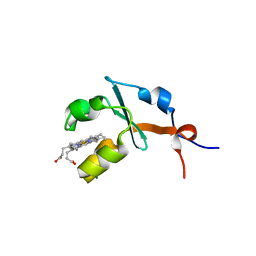 | | Structure of septuple mutant of Rat Outer Mitochondrial Membrane Cytochrome B5 | | Descriptor: | Cytochrome b5 type B, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Terzyan, S, Zhang, X.C, Benson, D.R, Wang, L, Sun, N. | | Deposit date: | 2006-09-01 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A histidine/tryptophan pi-stacking interaction stabilizes the heme-independent folding core of microsomal apocytochrome b5 relative to that of mitochondrial apocytochrome b5.
Biochemistry, 45, 2006
|
|
1BM6
 
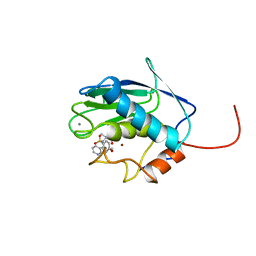 | | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN STROMELYSIN-1 COMPLEXED TO A POTENT NON-PEPTIDIC INHIBITOR, NMR, 20 STRUCTURES | | Descriptor: | 1-METHYLOXY-4-SULFONE-BENZENE, 3-METHYLPYRIDINE, CALCIUM ION, ... | | Authors: | Li, Y, Zhang, X, Melton, R, Ganu, V, Gonnella, N.C. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin-1 complexed to a potent, nonpeptidic inhibitor.
Biochemistry, 37, 1998
|
|
2OBR
 
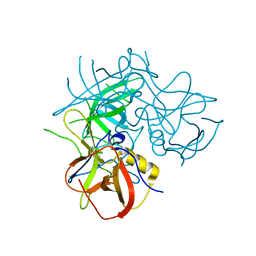 | | Crystal Structures of P Domain of Norovirus VA387 | | Descriptor: | Capsid protein | | Authors: | Cao, S, Lou, Z, Jiang, X, Zhang, X.C, Li, X, Rao, Z. | | Deposit date: | 2006-12-20 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of blood group trisaccharides by norovirus.
J.Virol., 81, 2007
|
|
4N8R
 
 | | Crystal structure of RXRa LBD complexed with a synthetic modulator K-8008 | | Descriptor: | 5-(2-{(1Z)-2-methyl-1-[4-(propan-2-yl)benzylidene]-1H-inden-3-yl}ethyl)-1H-tetrazole, Retinoic acid receptor RXR-alpha | | Authors: | Aleshin, A.E, Su, Y, Zhang, X, Liddington, R.C. | | Deposit date: | 2013-10-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Sulindac-Derived RXR alpha Modulators Inhibit Cancer Cell Growth by Binding to a Novel Site.
Chem.Biol., 21, 2014
|
|
6N9Y
 
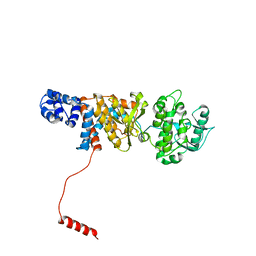 | | Atomic structure of Non-Structural protein 1 of bluetongue virus | | Descriptor: | Non-structural protein 1 | | Authors: | Kerviel, A, Ge, P, Lai, M, Jih, J, Boyce, M, Zhang, X, Zhou, Z.H, Roy, P. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic structure of the translation regulatory protein NS1 of bluetongue virus.
Nat Microbiol, 4, 2019
|
|
7BWA
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 2 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BW7
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin. | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-13 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BW8
 
 | | Cryo-EM Structure for the Insulin Binding Region in the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
4N5G
 
 | | Crystal Structure of RXRa LBD complexed with a synthetic modulator K8012 | | Descriptor: | 5-(2-{(1Z)-5-fluoro-2-methyl-1-[4-(propan-2-yl)benzylidene]-1H-inden-3-yl}ethyl)-1H-tetrazole, Retinoic acid receptor RXR-alpha | | Authors: | Aleshin, A.E, Su, Y, Zhang, X, Liddington, R.C. | | Deposit date: | 2013-10-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Sulindac-Derived RXR alpha Modulators Inhibit Cancer Cell Growth by Binding to a Novel Site.
Chem.Biol., 21, 2014
|
|
