6AK4
 
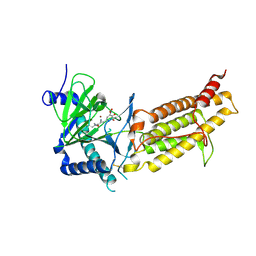 | | Crystal structure of human FTO in complex with small-molecule inhibitors | | Descriptor: | (~{E})-2-cyano-~{N},~{N}-diethyl-3-[3-nitro-4,5-bis(oxidanyl)phenyl]prop-2-enamide, Alpha-ketoglutarate-dependent dioxygenase FTO,Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | Authors: | Wang, Y, Cao, R, Peng, S, Zhang, W, Huang, N. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1.
Sci Transl Med, 11, 2019
|
|
2V79
 
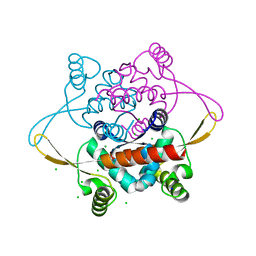 | | Crystal Structure of the N-terminal domain of DnaD from Bacillus Subtilis | | Descriptor: | CHLORIDE ION, DNA REPLICATION PROTEIN DNAD, SODIUM ION | | Authors: | Schneider, S, Zhang, W, Soultanas, P, Paoli, M. | | Deposit date: | 2007-07-27 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the N-Terminal Oligomerization Domain of Dnad Reveals a Unique Tetramerization Motif and Provides Insights Into Scaffold Formation.
J.Mol.Biol., 376, 2008
|
|
2Y2C
 
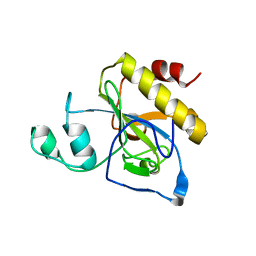 | | crystal structure of AmpD Apoenzyme | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y28
 
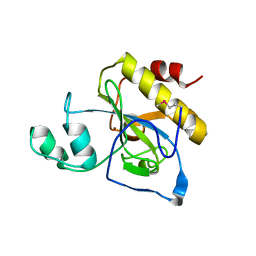 | | crystal structure of Se-Met AmpD derivative | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2B
 
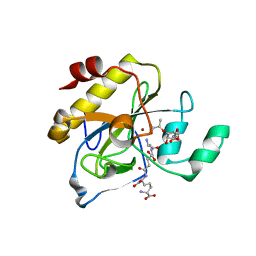 | | crystal structure of AmpD in complex with reaction products | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
1THD
 
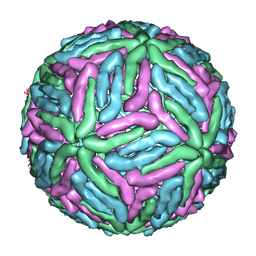 | | COMPLEX ORGANIZATION OF DENGUE VIRUS E PROTEIN AS REVEALED BY 9.5 ANGSTROM CRYO-EM RECONSTRUCTION | | Descriptor: | Major envelope protein E | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2004-06-01 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
2Y2E
 
 | | crystal structure of AmpD grown at pH 5.5 | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2D
 
 | | crystal structure of AmpD holoenzyme | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
4PXZ
 
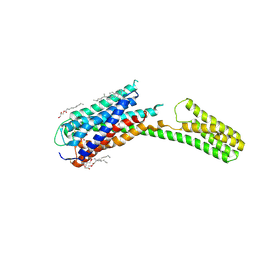 | | Crystal structure of P2Y12 receptor in complex with 2MeSADP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), CHOLESTEROL, ... | | Authors: | Zhang, J, Zhang, K, Gao, Z.G, Paoletta, S, Zhang, D, Han, G.W, Li, T, Ma, L, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Agonist-bound structure of the human P2Y12 receptor
Nature, 509, 2014
|
|
4PZA
 
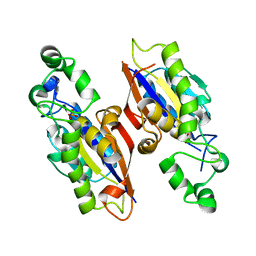 | | The complex structure of mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c with inorganic phosphate | | Descriptor: | Glucosyl-3-phosphoglycerate phosphatase, PHOSPHATE ION | | Authors: | Zhou, W.H, Zheng, Q.Q, Jiang, D.Q, Zhang, W, Zhang, Q.Q, Jin, J, Li, X, Yang, H.T, Shaw, N, Rao, Z. | | Deposit date: | 2014-03-29 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase
J.Biol.Chem., 289, 2014
|
|
4PZ9
 
 | | The native structure of mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c | | Descriptor: | Glucosyl-3-phosphoglycerate phosphatase | | Authors: | Zhou, W.H, Zheng, Q.Q, Jiang, D.Q, Zhang, W, Zhang, Q.Q, Jin, J, Li, X, Yang, H.T, Shaw, N, Rao, Z. | | Deposit date: | 2014-03-28 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase
J.Biol.Chem., 289, 2014
|
|
4QIH
 
 | | The structure of mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c complexes with VO3 | | Descriptor: | Glucosyl-3-phosphoglycerate phosphatase, VANADATE ION | | Authors: | Zhou, W.H, Zheng, Q.Q, Jiang, D.Q, Zhang, W, Zhang, Q.Q, Jin, J, Li, X, Yang, H.T, Shaw, N, Rao, Z. | | Deposit date: | 2014-05-30 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase
J.Biol.Chem., 289, 2014
|
|
4NTJ
 
 | | Structure of the human P2Y12 receptor in complex with an antithrombotic drug | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, P2Y purinoceptor 12,Soluble cytochrome b562,P2Y purinoceptor 12, ... | | Authors: | Zhang, K, Zhang, J, Gao, Z.-G, Zhang, D, Zhu, L, Han, G.W, Moss, S.M, Paoletta, S, Kiselev, E, Lu, W, Fenalti, G, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the human P2Y12 receptor in complex with an antithrombotic drug
Nature, 509, 2014
|
|
4MZ7
 
 | | Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Zhu, C, Gao, W, Zhao, K, Qin, X, Zhang, Y, Peng, X, Zhang, L, Dong, Y, Zhang, W, Li, P, Wei, W, Gong, Y, Yu, X.F. | | Deposit date: | 2013-09-29 | | Release date: | 2013-11-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase
Nat Commun, 4, 2013
|
|
6F89
 
 | | Structure of H234A/Y235A P.abyssi Sua5 | | Descriptor: | BICARBONATE ION, THREONINE, Threonylcarbamoyl-AMP synthase | | Authors: | Pichard-Kostuch, A, Zhang, W, Liger, D, Daugeron, M.C, Letoquart, J, Li de la Sierra-Gallay, I, Forterre, P, Collinet, B, van Tilbeurgh, H, Basta, T. | | Deposit date: | 2017-12-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure-function analysis of Sua5 protein reveals novel functional motifs required for the biosynthesis of the universal t6A tRNA modification.
RNA, 24, 2018
|
|
6F87
 
 | | Crystal structure of P. abyssi Sua5 complexed with L-threonine and PPi | | Descriptor: | PYROPHOSPHATE 2-, THREONINE, Threonylcarbamoyl-AMP synthase | | Authors: | Pichard-Kostuch, A, Zhang, W, Liger, D, Daugeron, M.C, Letoquart, J, Li de la Sierra-Gallay, I, Forterre, P, Collinet, B, van Tilbeurgh, H, Basta, T. | | Deposit date: | 2017-12-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure-function analysis of Sua5 protein reveals novel functional motifs required for the biosynthesis of the universal t6A tRNA modification.
RNA, 24, 2018
|
|
6F8Y
 
 | | Crystal structure of P. abyssi Sua5 complexed with L-threonine | | Descriptor: | THREONINE, Threonylcarbamoyl-AMP synthase | | Authors: | Pichard-Kostuch, A, Zhang, W, Liger, D, Daugeron, M.C, Letoquart, J, Li de la Sierra-Gallay, I, Forterre, P, Collinet, B, van Tilbeurgh, H, Basta, T. | | Deposit date: | 2017-12-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure-function analysis of Sua5 protein reveals novel functional motifs required for the biosynthesis of the universal t6A tRNA modification.
RNA, 24, 2018
|
|
6GX5
 
 | | Narrow Pick Filament from Pick's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zhang, W, Murzin, A.G, Murshudov, G, Garringer, H.J, Vidal, R, Crowther, R.A, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2018-06-26 | | Release date: | 2018-09-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of filaments from Pick's disease reveal a novel tau protein fold.
Nature, 561, 2018
|
|
1RQL
 
 | | Crystal Structure of Phosponoacetaldehyde Hydrolase Complexed with Magnesium and the Inhibitor Vinyl Sulfonate | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase, VINYLSULPHONIC ACID | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
1RQN
 
 | | Phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
1TGE
 
 | | The structure of immature Dengue virus at 12.5 angstrom | | Descriptor: | envelope glycoprotein | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2004-05-28 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein.
Structure, 12, 2004
|
|
1SWV
 
 | | Crystal structure of the D12A mutant of phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
1TG8
 
 | | The structure of Dengue virus E glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, envelope glycoprotein | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Rossmann, M.G. | | Deposit date: | 2004-05-28 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
1SWW
 
 | | Crystal structure of the phosphonoacetaldehyde hydrolase D12A mutant complexed with magnesium and substrate phosphonoacetaldehyde | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETALDEHYDE, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
1HOC
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF H-2DB AT 2.4 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ANTIGEN-DETERMINANT SELECTION | | Descriptor: | 9-RESIDUE PEPTIDE, BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (H2-DB) (ALPHA CHAIN) | | Authors: | Young, A.C.M, Zhang, W, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1994-04-30 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of H-2Db at 2.4 A resolution: implications for antigen-determinant selection
Cell(Cambridge,Mass.), 76, 1994
|
|
