3MFQ
 
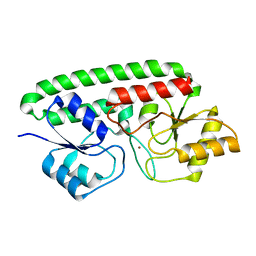 | | A Glance into the Metal Binding Specificity of TroA: Where Elaborate Behaviors Occur in the Active Center | | Descriptor: | High-affinity zinc uptake system protein znuA, ZINC ION | | Authors: | Gao, G.F, Zheng, B, Zhang, Q, Gao, J, Han, H, Li, M. | | Deposit date: | 2010-04-03 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Insight into the interaction of metal ions with TroA from Streptococcus suis
Plos One, 6, 2011
|
|
1SH4
 
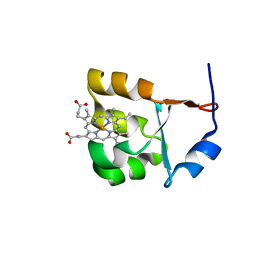 | |
1J0Q
 
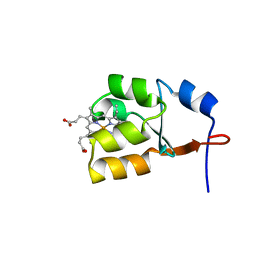 | | Solution Structure of Oxidized Bovine Microsomal Cytochrome b5 mutant V61H | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Wu, H, Huang, Z, Cao, C, Zhang, Q, Wang, Y.-H, Ma, J.-B, Xue, L.-L. | | Deposit date: | 2002-11-20 | | Release date: | 2003-08-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the oxidized bovine microsomal cytochrome b5 mutant V61H
Biochem.Biophys.Res.Commun., 307, 2003
|
|
1WWN
 
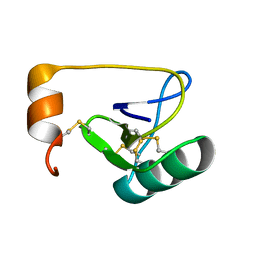 | | NMR Solution Structure of BmK-betaIT, an Excitatory Scorpion Toxin from Buthus martensi Karsch | | Descriptor: | Excitatory insect selective toxin 1 | | Authors: | Wu, H, Tong, X, Chen, X, Zhang, Q, Zheng, X, Zhang, N, Wu, G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of BmK-betaIT, an excitatory scorpion beta-toxin without a 'hot spot' at the relevant position
Biochem.Biophys.Res.Commun., 349, 2006
|
|
1SZV
 
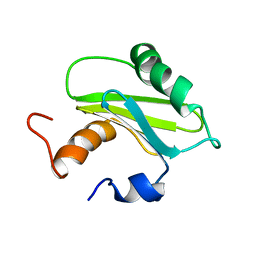 | | Structure of the Adaptor Protein p14 reveals a Profilin-like Fold with Novel Function | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein | | Authors: | Qian, C, Zhang, Q, Wang, X, Zeng, L, Farooq, A, Zhou, M.M. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Adaptor Protein p14 Reveals a Profilin-like Fold with Distinct Function
J.Mol.Biol., 347, 2005
|
|
1SJ6
 
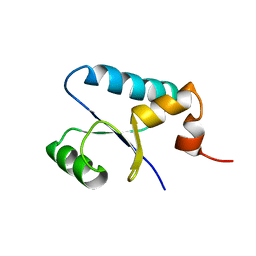 | | NMR Structure and Regulated Expression in APL Cell of Human SH3BGRL3 | | Descriptor: | SH3 domain-binding glutamic acid-rich-like protein 3 | | Authors: | Xu, C, Tang, Y, Xu, Y, Wu, J, Shi, Y, Zhang, Q, Zheng, P, Du, Y. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure and regulated expression in APL cell of human SH3BGRL3.
Febs Lett., 579, 2005
|
|
6A96
 
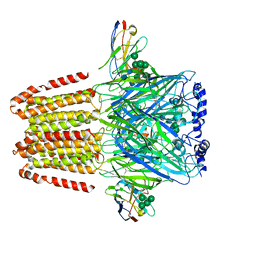 | | Cryo-EM structure of the human alpha5beta3 GABAA receptor in complex with GABA and Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-5,Gamma-aminobutyric acid receptor subunit alpha-5, ... | | Authors: | Liu, S, Xu, L, Guan, F, Liu, Y.T, Cui, Y, Zhang, Q, Bi, G.Q, Zhou, Z.H, Zhang, X, Ye, S. | | Deposit date: | 2018-07-11 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM structure of the human alpha 5 beta 3 GABAAreceptor.
Cell Res., 28, 2018
|
|
5IWW
 
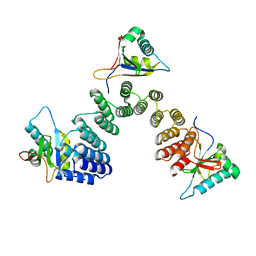 | | Crystal structure of RNA editing factor of designer PLS-type PPR/9R protein in complex with MORF9/RIP9 | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic, PLS9-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
5C4A
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Saphr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5C4X
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5C3E
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5C4J
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5IZW
 
 | | Crystal structure of RNA editing specific factor of designer PLS-type PPR-9R protein | | Descriptor: | PLS9-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-26 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
5H9M
 
 | | Crystal structure of siah2 SBD domain | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase SIAH2, PENTAETHYLENE GLYCOL, ... | | Authors: | Dong, A, Zhang, Q, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Crystal structure of siah2 SBD domain
to be published
|
|
4B03
 
 | | 6A Electron cryomicroscopy structure of immature Dengue virus serotype 1 | | Descriptor: | DENGUE VIRUS 1 E PROTEIN, DENGUE VIRUS 1 PRM PROTEIN | | Authors: | Kostyuchenko, V.A, Zhang, Q, Tan, L.C, Ng, T.S, Lok, S.M. | | Deposit date: | 2012-06-28 | | Release date: | 2013-06-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Immature and Mature Dengue Serotype 1 Virus Structures Provide Insight Into the Maturation Process.
J.Virol., 87, 2013
|
|
5YK1
 
 | | The complex structure of Rv3197-AMPPNP from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable conserved ATP-binding protein ABC transporter, ... | | Authors: | Rao, Z.H, Zhang, Q.Q. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Discovery of the first macrolide antibiotic binding protein in Mycobacterium tuberculosis: a new antibiotic resistance drug target.
Protein Cell, 9, 2018
|
|
5YK0
 
 | | The complex structure of Rv3197-ADP from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Probable conserved ATP-binding protein ABC transporter, ... | | Authors: | Rao, Z.H, Zhang, Q.Q. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Discovery of the first macrolide antibiotic binding protein in Mycobacterium tuberculosis: a new antibiotic resistance drug target.
Protein Cell, 9, 2018
|
|
5YK2
 
 | |
4Z30
 
 | | Crystal structure of the ROQ domain of human Roquin-2 | | Descriptor: | Roquin-2, UNKNOWN ATOM OR ION | | Authors: | DONG, A, ZHANG, Q, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-30 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | New Insights into the RNA-Binding and E3 Ubiquitin Ligase Activities of Roquins.
Sci Rep, 5, 2015
|
|
4Z31
 
 | | Crystal structure of the RC3H2 ROQ domain in complex with stem-loop and double-stranded forms of RNA | | Descriptor: | CHLORIDE ION, RNA (5'-R(*A)-D(P*UP*GP*UP*UP*CP*UP*GP*UP*GP*AP*AP*CP*AP*C)-3'), Roquin-2, ... | | Authors: | DONG, A, ZHANG, Q, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-30 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New Insights into the RNA-Binding and E3 Ubiquitin Ligase Activities of Roquins.
Sci Rep, 5, 2015
|
|
5C44
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
6DNY
 
 | |
6BNH
 
 | | Solution NMR structures of BRD4 ET domain with JMJD6 peptide | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, Bromodomain-containing protein 4 | | Authors: | Konuma, T, Yu, D, Zhao, C, Ju, Y, Sharma, R, Ren, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4.
Sci Rep, 7, 2017
|
|
3JBM
 
 | | Electron cryo-microscopy of a virus-like particle of orange-spotted grouper nervous necrosis virus | | Descriptor: | virus-like particle of orange-spotted grouper nervous necrosis virus | | Authors: | Xie, J, Li, K, Gao, Y, Huang, R, Lai, Y, Shi, Y, Yang, S, Zhu, G, Zhang, Q, He, J. | | Deposit date: | 2015-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis and insertion study reveal the ideal sites for surface displaying foreign peptides on a betanodavirus-like particle
Vet. Res., 47, 2016
|
|
3K9V
 
 | | Crystal structure of rat mitochondrial P450 24A1 S57D in complex with CHAPS | | Descriptor: | 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ... | | Authors: | Annalora, A.J, Goodin, D.B, Hong, W, Zhang, Q, Johnson, E.F, Stout, C.D. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism.
J.Mol.Biol., 396, 2010
|
|
