5F3Y
 
 | | Crystal Structure of Myo7b N-MyTH4-FERM-SH3 in complex with Anks4b CEN | | Descriptor: | Ankyrin repeat and SAM domain-containing protein 4B, Unconventional myosin-VIIb | | Authors: | Li, J, He, Y, Lu, Q, Zhang, M. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.409 Å) | | Cite: | Mechanistic Basis of Organization of the Harmonin/USH1C-Mediated Brush Border Microvilli Tip-Link Complex
Dev.Cell, 36, 2016
|
|
5F67
 
 | | An exquisitely specific PDZ/target recognition revealed by the structure of INAD PDZ3 in complex with TRP channel tail | | Descriptor: | Inactivation-no-after-potential D protein, TRP C terminal Tail | | Authors: | Ye, F, Shang, Y, Liu, W, Zhang, M. | | Deposit date: | 2015-12-05 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An Exquisitely Specific PDZ/Target Recognition Revealed by the Structure of INAD PDZ3 in Complex with TRP Channel Tail
Structure, 24, 2016
|
|
5YPR
 
 | | Crystal Structure of PSD-95 SH3-GK domain in complex with a synthesized inhibitor | | Descriptor: | Disks large homolog 4, Synthesized GK inhibitor | | Authors: | Zhu, J, Zhou, Q, Shang, Y, Weng, Z, Zhu, R, Zhang, M. | | Deposit date: | 2017-11-02 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Synaptic Targeting and Function of SAPAPs Mediated by Phosphorylation-Dependent Binding to PSD-95 MAGUKs.
Cell Rep, 21, 2017
|
|
4YTK
 
 | | Structure of the KOW1-Linker1 domain of Transcription Elongation Factor Spt5 | | Descriptor: | Transcription elongation factor SPT5 | | Authors: | Meyer, P.A, Li, S, Zhang, M, Yamada, K, Takagi, Y, Hartzog, G.A, Fu, J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.0904 Å) | | Cite: | Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5.
Mol.Cell.Biol., 35, 2015
|
|
4YTL
 
 | | Structure of the KOW2-KOW3 Domain of Transcription Elongation Factor Spt5. | | Descriptor: | GLYCEROL, SULFATE ION, Transcription elongation factor SPT5 | | Authors: | Meyer, P.A, Li, S, ZHang, M, Yamada, K, Takagi, Y, Hartzog, G.A, Fu, J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5.
Mol.Cell.Biol., 35, 2015
|
|
4YL8
 
 | | Crystal structure of the Crumbs/Moesin complex | | Descriptor: | GLYCEROL, IODIDE ION, Moesin, ... | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Phosphorylation-regulated Interaction between the Cytoplasmic Tail of Cell Polarity Protein Crumbs and the Actin-binding Protein Moesin
J.Biol.Chem., 290, 2015
|
|
4ZRK
 
 | | Merlin-FERM and Lats1 complex | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS1 | | Authors: | Lin, Z, Li, Y, Wei, Z, Zhang, M. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4Q32
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and C91 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(naphthalen-2-yl)-2-[2-(pyridin-2-yl)-1H-benzimidazol-1-yl]acetamide | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Gu, M, Zhang, M, Mandapati, K, Gollapalli, D.R, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and C91
To be Published
|
|
3HPK
 
 | |
4Q33
 
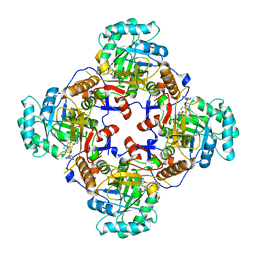 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and A110 | | Descriptor: | 4-[(1R)-1-[1-(4-chlorophenyl)-1,2,3-triazol-4-yl]ethoxy]-1-oxidanyl-quinoline, ACETIC ACID, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Gu, M, Zhang, M, Mandapati, K, Gollapalli, D.R, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.885 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and A110
TO BE PUBLISHED
|
|
4QM1
 
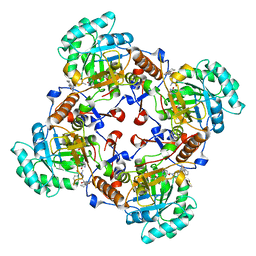 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor D67 | | Descriptor: | 2-(3-methyl-4-oxo-3,4-dihydrophthalazin-1-yl)-N-(6,7,8,9-tetrahydrodibenzo[b,d]furan-2-yl)acetamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Mandapati, K, Gollapalli, D, Gorla, S.K, Zhang, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-14 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7964 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor D67
To be Published, 2014
|
|
3K1R
 
 | |
3H8D
 
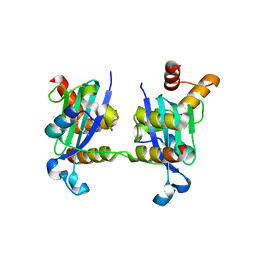 | | Crystal structure of Myosin VI in complex with Dab2 peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, Disabled homolog 2, ... | | Authors: | Yu, C, Feng, W, Wei, Z, Zhang, M. | | Deposit date: | 2009-04-29 | | Release date: | 2009-09-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Myosin VI undergoes cargo-mediated dimerization
Cell(Cambridge,Mass.), 138, 2009
|
|
3G5B
 
 | | The structure of UNC5b cytoplasmic domain | | Descriptor: | Netrin receptor UNC5B, PHOSPHATE ION | | Authors: | Wang, R, Wei, Z, Zhang, M. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Autoinhibition of UNC5b revealed by the cytoplasmic domain structure of the receptor
Mol.Cell, 33, 2009
|
|
3HPM
 
 | |
4HWW
 
 | | Crystal structure of human Arginase-1 complexed with inhibitor 9 | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5R)-5-amino-5-carboxy-7-(piperidin-1-yl)heptyl](trihydroxy)borate(1-) | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D.L, Golebiowski, A, Ji, M, Zhang, M, Beckett, P, Sheeler, R, Andreoli, M, Conway, B, Mahboubi, K, Schroeter, H, Van Zandt, M.C, Podjarny, A. | | Deposit date: | 2012-11-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Discovery of (R)-2-Amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic Acid and Congeners As Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury.
J.Med.Chem., 56, 2013
|
|
3SBJ
 
 | | MutM slanted complex 7 | | Descriptor: | 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*AP*GP*G)-3', 5'-D(P*CP*CP*TP*GP*GP*TP*(CX)P*TP*AP*CP*C)-3', Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Verdine, G.L. | | Deposit date: | 2011-06-05 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Strandwise translocation of a DNA glycosylase on undamaged DNA.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4I06
 
 | | Crystal structure of human Arginase-2 complexed with inhibitor 14 | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D.L, Golebiowski, A, Ji, M, Zhang, M, Beckett, P, Sheeler, R, Andreoli, M, Conway, B, Mahboubi, K, Schroeter, H, Van Zandt, M.C, Podjarny, A. | | Deposit date: | 2012-11-16 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of (R)-2-Amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic Acid and Congeners As Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury.
J.Med.Chem., 56, 2013
|
|
4NQ0
 
 | | Structural insights into yeast histone chaperone Hif1: a scaffold protein recruiting protein complexes to core histones | | Descriptor: | HAT1-interacting factor 1 | | Authors: | Liu, H, Zhang, M, He, W, Zhu, Z, Teng, M, Gao, Y, Niu, L. | | Deposit date: | 2013-11-23 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into yeast histone chaperone Hif1: a scaffold protein recruiting protein complexes to core histones
Biochem.J., 462, 2014
|
|
5YIS
 
 | | Crystal Structure of AnkB LIR/LC3B complex | | Descriptor: | Ankyrin-2, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3B, ... | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
5YIR
 
 | | Crystal Structure of AnkB LIR/GABARAP complex | | Descriptor: | Ankyrin-2, Gamma-aminobutyric acid receptor-associated protein, NICKEL (II) ION | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
5YIQ
 
 | | Crystal structure of AnkG LIR/LC3B complex | | Descriptor: | Ankyrin-3, Microtubule-associated proteins 1A/1B light chain 3B, ZINC ION | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
4FYP
 
 | | Crystal Structure of Plant Vegetative Storage Protein | | Descriptor: | MAGNESIUM ION, Vegetative storage protein 1 | | Authors: | Chen, Y, Wei, J, Wang, M, Gong, W, Zhang, M. | | Deposit date: | 2012-07-05 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Arabidopsis VSP1 reveals the plant class C-like phosphatase structure of the DDDD superfamily of phosphohydrolases
Plos One, 7, 2012
|
|
4G5R
 
 | | Structure of LGN GL4/Galphai3 complex | | Descriptor: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | Deposit date: | 2012-07-18 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.481 Å) | | Cite: | Crystal Structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from Galphai subunits in G-coupled heterotrimeric proteins
To be Published
|
|
5YIP
 
 | | Crystal Structure of AnkG LIR/GABARAPL1 complex | | Descriptor: | Ankyrin-3, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1 | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
