6IRC
 
 | | C-terminal domain of Drosophila phospholipase b NORPA, methylated | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase | | Authors: | Ye, F, Li, J, Huang, Y, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.538 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
6IRB
 
 | | C-terminal coiled coil domain of Drosophila phospholipase C beta NORPA, selenomethionine | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase | | Authors: | Ye, F, Li, J, Huang, Y, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.661 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
6J69
 
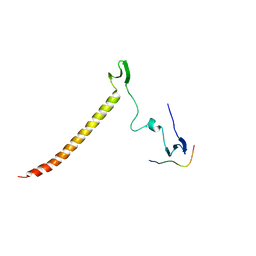 | | Structure of KIBRA and Dendrin Complex | | Descriptor: | Peptide from Dendrin, Protein KIBRA | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Kibra Modulates Learning and Memory via Binding to Dendrin.
Cell Rep, 26, 2019
|
|
6KG7
 
 | | Cryo-EM Structure of the Mammalian Tactile Channel Piezo2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Piezo-type mechanosensitive ion channel component 2 | | Authors: | Wang, L, Zhou, H, Zhang, M, Liu, W, Deng, T, Zhao, Q, Li, Y, Lei, J, Li, X, Xiao, B. | | Deposit date: | 2019-07-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and mechanogating of the mammalian tactile channel PIEZO2.
Nature, 573, 2019
|
|
6M3Q
 
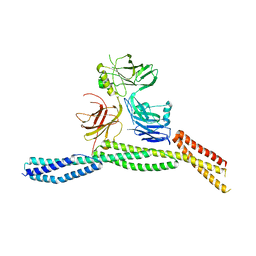 | | Crystal structure of AnkB/beta4-spectrin complex | | Descriptor: | Ankyrin-2, Spectrin beta chain | | Authors: | Li, J, Chen, K, Zhu, R, Zhang, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.436 Å) | | Cite: | Structural Basis Underlying Strong Interactions between Ankyrins and Spectrins.
J.Mol.Biol., 432, 2020
|
|
6LNM
 
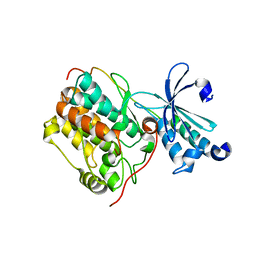 | | Crystal structure of CASK-CaMK in complex with Mint1-CID | | Descriptor: | Amyloid-beta A4 precursor protein-binding family A member 1, Peripheral plasma membrane protein CASK | | Authors: | Cai, Q, Zhang, M. | | Deposit date: | 2019-12-31 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the High-Affinity Interaction between CASK and Mint1.
Structure, 28, 2020
|
|
6M3R
 
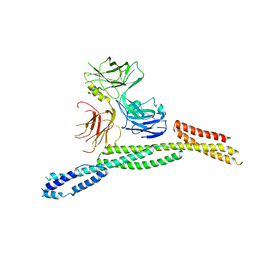 | | Crystal structure of AnkG/beta4-spectrin complex | | Descriptor: | Ankyrin-3, Spectrin beta chain | | Authors: | Li, J, Chen, K, Zhu, R, Zhang, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.313 Å) | | Cite: | Structural Basis Underlying Strong Interactions between Ankyrins and Spectrins.
J.Mol.Biol., 432, 2020
|
|
7CHA
 
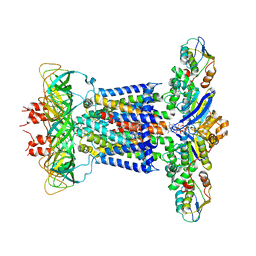 | | Cryo-EM structure of P.aeruginosa MlaFEBD with AMPPNP | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, MlaD domain-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7YUY
 
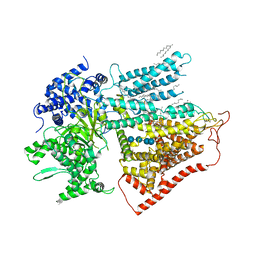 | | Structure of a mutated membrane-bound glycosyltransferase | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | Deposit date: | 2022-08-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
7CH6
 
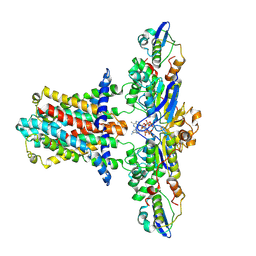 | | Cryo-EM structure of E.coli MlaFEB with AMPPNP | | Descriptor: | Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CH8
 
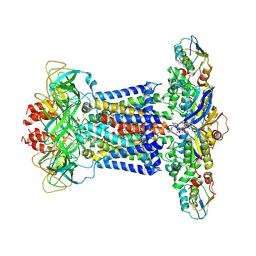 | | Cryo-EM structure of P.aeruginosa MlaFEBD with ADP-V | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CH9
 
 | | Cryo-EM structure of P.aeruginosa MlaFEBD | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, MlaD domain-containing protein, Probable ATP-binding component of ABC transporter, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
8V2G
 
 | |
8V2H
 
 | |
6LYW
 
 | | Structural insight into the biological functions of Arabidopsis thaliana ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
6LYX
 
 | | Crystal structure of oxidized ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Cai, W.G, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
2HQ4
 
 | | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH1570 | | Authors: | Li, Y, Marshall, M, Chang, J, Zhao, M, Zhang, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii
To be Published
|
|
5ET0
 
 | | Crystal structure of Myo3b-ARB2 in complex with Espin1-AR | | Descriptor: | Espin, Myosin-IIIb | | Authors: | Liu, H, Li, J, Liu, W, Zhang, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Myosin III-mediated cross-linking and stimulation of actin bundling activity of Espin
Elife, 5, 2016
|
|
5ET1
 
 | | Crystal structure of Myo3b-ARB1 in complex with Espin1-AR | | Descriptor: | Espin, GLYCEROL, Myosin-IIIb | | Authors: | Liu, H, Li, J, liu, W, Zhang, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Myosin III-mediated cross-linking and stimulation of actin bundling activity of Espin
Elife, 5, 2016
|
|
5F3X
 
 | | Crystal structure of Harmonin NPDZ1 in complex with ANKS4B SAM-PBM | | Descriptor: | Ankyrin repeat and SAM domain-containing protein 4B, CHLORIDE ION, Harmonin | | Authors: | Li, J, He, Y, Lu, Q, Zhang, M. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Mechanistic Basis of Organization of the Harmonin/USH1C-Mediated Brush Border Microvilli Tip-Link Complex
Dev.Cell, 36, 2016
|
|
5F3Y
 
 | | Crystal Structure of Myo7b N-MyTH4-FERM-SH3 in complex with Anks4b CEN | | Descriptor: | Ankyrin repeat and SAM domain-containing protein 4B, Unconventional myosin-VIIb | | Authors: | Li, J, He, Y, Lu, Q, Zhang, M. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.409 Å) | | Cite: | Mechanistic Basis of Organization of the Harmonin/USH1C-Mediated Brush Border Microvilli Tip-Link Complex
Dev.Cell, 36, 2016
|
|
5F67
 
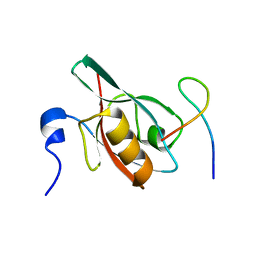 | | An exquisitely specific PDZ/target recognition revealed by the structure of INAD PDZ3 in complex with TRP channel tail | | Descriptor: | Inactivation-no-after-potential D protein, TRP C terminal Tail | | Authors: | Ye, F, Shang, Y, Liu, W, Zhang, M. | | Deposit date: | 2015-12-05 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An Exquisitely Specific PDZ/Target Recognition Revealed by the Structure of INAD PDZ3 in Complex with TRP Channel Tail
Structure, 24, 2016
|
|
8I3E
 
 | |
2B8K
 
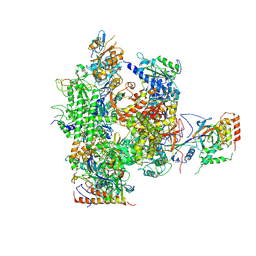 | | 12-subunit RNA Polymerase II | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, DNA-directed RNA polymerase II 19 kDa polypeptide, ... | | Authors: | Meyer, P.A, Ye, P, Zhang, M, Suh, M.H, Fu, J. | | Deposit date: | 2005-10-07 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Phasing RNA Polymerase II Using Intrinsically Bound Zn Atoms: An Updated Structural Model.
Structure, 14, 2006
|
|
2FJL
 
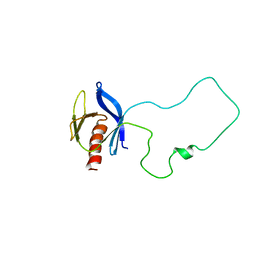 | | Solution Structure of the Split PH domain in Phospholipase C-gamma1 | | Descriptor: | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2006-01-03 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Split Pleckstrin Homology Domain in Phospholipase C-{gamma}1 and Its Interaction with TRPC3
J.Biol.Chem., 281, 2006
|
|
