8IOB
 
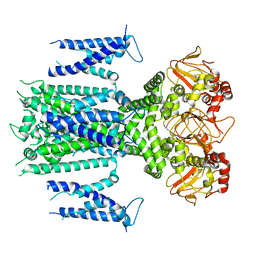 | | Herg1a-herg1b closed state 1 | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of Herg1a-herg1b closed state 1
To Be Published
|
|
8IO4
 
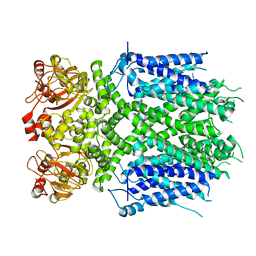 | | Herg1a-herg1b open state | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Herg1a-herg1b open state
To Be Published
|
|
5PNT
 
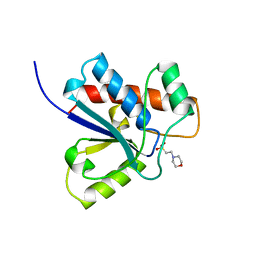 | | CRYSTAL STRUCTURE OF A HUMAN LOW MOLECULAR WEIGHT PHOSPHOTYROSYL PHOSPHATASE. IMPLICATIONS FOR SUBSTRATE SPECIFICITY | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LOW MOLECULAR WEIGHT PHOSPHOTYROSYL PHOSPHATASE | | Authors: | Zhang, M, Stauffacher, C, Lin, D, Vanetten, R. | | Deposit date: | 1998-04-29 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a human low molecular weight phosphotyrosyl phosphatase. Implications for substrate specificity.
J.Biol.Chem., 273, 1998
|
|
6A6J
 
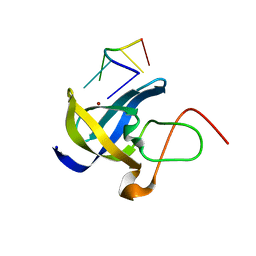 | | Crystal structure of Zebra fish Y-box protein1 (YB-1) Cold-shock domain in complex with 6mer m5C RNA | | Descriptor: | RNA (5'-R(P*CP*AP*UP*(5MC)P*U)-3'), ZINC ION, Zebra fish Y-box protein1 (YB-1) | | Authors: | Zhang, M.M, Wu, B.X, Huang, Y, Ma, J.B. | | Deposit date: | 2018-06-28 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay.
Mol.Cell, 75, 2019
|
|
7L0R
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0P
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0S
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LH9
 
 | | Crystal structure of BRPF2 PWWP domain in complex with DNA | | Descriptor: | Bromodomain-containing protein 1, DNA | | Authors: | Zhang, M, Lei, M, Qin, S, Dong, A, Yang, A, Li, Y, Loppnau, P, Hughes, T.R, Arrowsmith, C.H, Edwards, A.M, Min, J, Liu, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-21 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the BRPF2 PWWP domain in complex with DNA reveals a different binding mode than the HDGF family of PWWP domains.
Biochim Biophys Acta Gene Regul Mech, 1864, 2021
|
|
8OOS
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-hexasome refinement state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase Ino80, DNA Strand 2, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
7L0Q
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6IMC
 
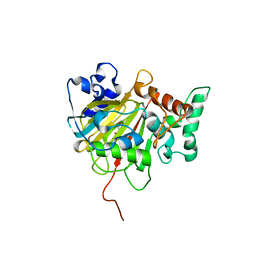 | | Crystal Structure of ALKBH1 in complex with Mn(II) and N-Oxalylglycine | | Descriptor: | MANGANESE (II) ION, N-OXALYLGLYCINE, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
7YFG
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (major class in asymmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFH
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine, glutamate and (R)-PYD-106 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YJ0
 
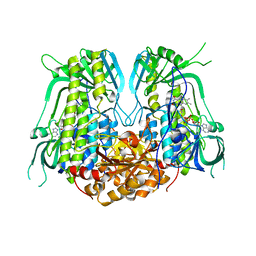 | |
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6JQF
 
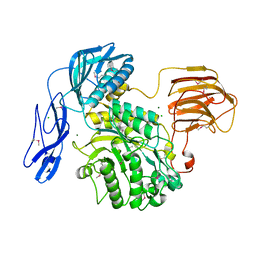 | |
5YUH
 
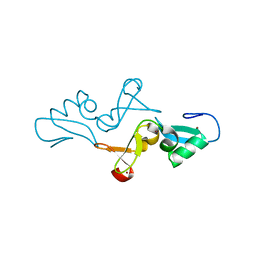 | |
5YUG
 
 | | AtVAL1 PHD-Like domain in the P31 space group | | Descriptor: | B3 domain-containing transcription repressor VAL1, GLYCEROL, ZINC ION | | Authors: | Zhang, M.M, Wu, B.X. | | Deposit date: | 2017-11-22 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | AtVAL1 PHD-Like domain in the P31 space group
To Be Published
|
|
2C2V
 
 | | Crystal structure of the CHIP-UBC13-UEV1a complex | | Descriptor: | STIP1 homology and U box-containing protein 1, Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 1 | | Authors: | Zhang, M, Roe, S.M, Pearl, L.H. | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex.
Mol. Cell, 20, 2005
|
|
8W6A
 
 | |
5XEC
 
 | | Heterodimer constructed from PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
2XCM
 
 | | COMPLEX OF HSP90 N-TERMINAL, SGT1 CS AND RAR1 CHORD2 DOMAIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CYTOSOLIC HEAT SHOCK PROTEIN 90, MAGNESIUM ION, ... | | Authors: | Zhang, M, Pearl, L.H. | | Deposit date: | 2010-04-23 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Assembly of Hsp90-Sgt1-Chord Protein Complexes: Implications for Chaperoning of Nlr Innate Immunity Receptors
Mol.Cell, 39, 2010
|
|
8WUL
 
 | | Crystal structure of affinity enhanced TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen, ... | | Authors: | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
2C2L
 
 | | Crystal structure of the CHIP U-box E3 ubiquitin ligase | | Descriptor: | CARBOXY TERMINUS OF HSP70-INTERACTING PROTEIN, HSP90, NICKEL (II) ION, ... | | Authors: | Zhang, M, Roe, S.M, Pearl, L.H. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Chaperoned Ubiquitylation-Crystal Structures of the Chip U Box E3 Ubiquitin Ligase and a Chip-Ubc13-Uev1A Complex
Mol.Cell, 20, 2005
|
|
5XED
 
 | | Heterodimer constructed from M61A PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
