5C1Y
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound 1 | | Descriptor: | 3C proteinase, propan-2-yl N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5C20
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound 2 | | Descriptor: | 2-methylpropyl N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
7BVC
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVE
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, Ethambutol, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVG
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
6KXW
 
 | | Crystal structure of human aquaporin AQP7 in bound to glycerol | | Descriptor: | Aquaporin-7, GLYCEROL | | Authors: | Zhang, L, Yao, D, Zhou, F, Zhang, Q, Zhou, L, Cao, Y. | | Deposit date: | 2019-09-13 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The structural basis for glycerol permeation by human AQP7
Sci Bull (Beijing), 66, 2020
|
|
8B35
 
 | |
8B32
 
 | |
8B3C
 
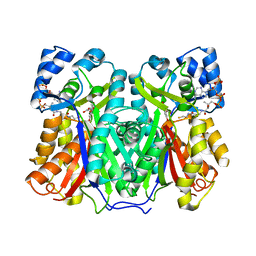 | | Chalcone synthase from Hordeum vulgare complexed with CoA and eriodictyol | | Descriptor: | (2S)-2-(3,4-DIHYDROXYPHENYL)-5,7-DIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, COENZYME A, Chalcone synthase 2 | | Authors: | Zhang, L, Groves, M.R. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a Plant Polyketide Synthase for the Biosynthesis of Methylated Flavonoids.
J.Agric.Food Chem., 72, 2024
|
|
8C9T
 
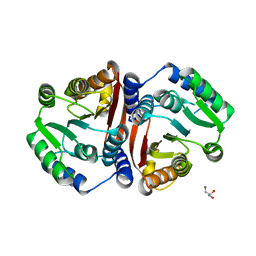 | | Catechol O-methyltransferase from Streptomyces avermitilis | | Descriptor: | GLYCEROL, Putative O-methyltransferase | | Authors: | Zhang, L, Groves, M.R. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Characterization and Extended Substrate Scope Analysis of Two Mg 2+ -Dependent O-Methyltransferases from Bacteria.
Chembiochem, 24, 2023
|
|
8C9V
 
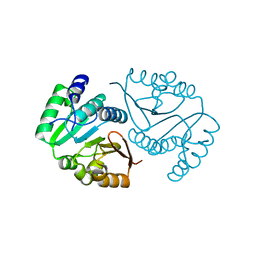 | | O-methyltransferase from Desulfuromonas acetoxidans | | Descriptor: | MAGNESIUM ION, O-methyltransferase, family 3 | | Authors: | Zhang, L, Groves, M.R. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Characterization and Extended Substrate Scope Analysis of Two Mg 2+ -Dependent O-Methyltransferases from Bacteria.
Chembiochem, 24, 2023
|
|
8C9S
 
 | |
6KXH
 
 | | Alp1U_Y247F mutant in complex with Fluostatin C | | Descriptor: | D-MALATE, Fluostatin C, Putative hydrolase, ... | | Authors: | Zhang, L, Yingli, Z, De, B.C, Zhang, C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78039551 Å) | | Cite: | Mutation of an atypical oxirane oxyanion hole improves regioselectivity of the alpha / beta-fold epoxide hydrolase Alp1U.
J.Biol.Chem., 295, 2020
|
|
6KXR
 
 | | Crystal structure of wild type Alp1U from the biosynthesis of kinamycins | | Descriptor: | D-MALATE, Putative hydrolase | | Authors: | Zhang, L, Yingli, Z, De, B.C, Zhang, C. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45073676 Å) | | Cite: | Mutation of an atypical oxirane oxyanion hole improves regioselectivity of the alpha / beta-fold epoxide hydrolase Alp1U.
J.Biol.Chem., 295, 2020
|
|
6Z2E
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with the activity-based probe, biotin-PEG(4)-Abu-Tle-Leu-Gln-vinylsulfone | | Descriptor: | (4~{S})-4-[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[3-[2-[2-[2-[2-[5-[(3~{a}~{S},4~{R},6~{a}~{R})-2-oxidanylidene-3,3~{a},4,6~{a}-tetrahydro-1~{H}-thieno[3,4-d]imidazol-4-yl]pentanoylamino]ethoxy]ethoxy]ethoxy]ethoxy]propanoylamino]butanoyl]amino]-3,3-dimethyl-butanoyl]amino]-4-methyl-pentanoyl]amino]-6-methylsulfonyl-hexanamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 Mpro in complex with the activity-based probe, biotin-PEG(4)-Abu-Tle-Leu-Gln-vinylsulfone
To Be Published
|
|
5XD6
 
 | | CARK1 phosphorylates ABA receptors | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein kinase superfamily protein | | Authors: | Zhang, L, Lou, Z. | | Deposit date: | 2017-03-27 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | CARK1 phosphorylates ABA receptors
To Be Published
|
|
6KVM
 
 | | Crystal structure of Chicken MHC Class II for 1.9 angstrom | | Descriptor: | GLYCEROL, MHC class II alpha chain, MHC class II beta chain 2, ... | | Authors: | Zhang, L, Xia, C. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | A Newly Recognized Pairing Mechanism of the alpha- and beta-Chains of the Chicken Peptide-MHC Class II Complex.
J Immunol., 204, 2020
|
|
6JV5
 
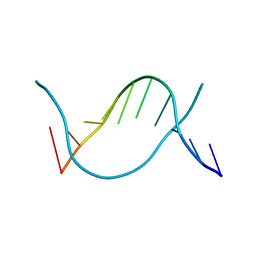 | | Crystal structure of 5-methylcytosine containing decamer dsDNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*(5CM)P*GP*CP*TP*GP*G)-3') | | Authors: | Zhang, L, Wang, Y.X. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
1KXK
 
 | |
7OQ6
 
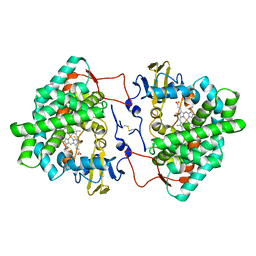 | | Crystal structure of cytochrome P450 Sas16 from Streptomyces asterosporus | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | Authors: | Zhang, L, Zhang, S, Bechthold, A, Einsle, O. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | P450-mediated dehydrotyrosine formation during WS9326 biosynthesis proceeds via dehydrogenation of a specific acylated dipeptide substrate.
Acta Pharm Sin B, 13, 2023
|
|
4ZJB
 
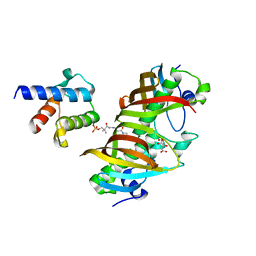 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) in complex with holo-ACP from Helicobacter pylori | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ, 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, ... | | Authors: | Zhang, L, Zhang, L, Shen, X, Jiang, H. | | Deposit date: | 2015-04-29 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of FabZ-ACP complex reveals a dynamic seesaw-like catalytic mechanism of dehydratase in fatty acid biosynthesis.
Cell Res., 26, 2016
|
|
6FV1
 
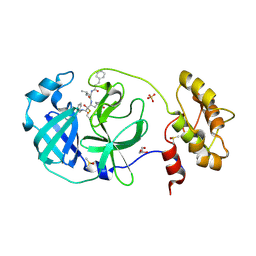 | | Structure of human coronavirus NL63 main protease in complex with the alpha-ketoamide (S)-N-((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)-2-cinnamamido-4-methylpentanamide (cinnamoyl-leucine-GlnLactam-CO-CO-NH-benzyl) | | Descriptor: | (2~{S})-4-methyl-~{N}-[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alpha-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication Structure-based design, synthesis, and activity assessment.
J.Med.Chem., 2020
|
|
6FV2
 
 | |
3JBL
 
 | | Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated Polymerization | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Zhang, L, Chen, S, Ruan, J, Wu, J, Tong, A.B, Yin, Q, Li, Y, David, L, Lu, A, Wang, W.L, Marks, C, Ouyang, Q, Zhang, X, Mao, Y, Wu, H. | | Deposit date: | 2015-09-05 | | Release date: | 2015-10-21 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization.
Science, 350, 2015
|
|
