7D8E
 
 | | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase in complex with LSB-09 | | Descriptor: | 1,2-ETHANEDIOL, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Dileep, K.V, Ihara, K, Sakai, N, Shirozu, M, Zhang, K.Y.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase in complex with LSB-09
To Be Published
|
|
7FIE
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-07-31 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FIZ
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-08-01 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FID
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon proteasecomplex (conformation 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-07-31 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
8J5Y
 
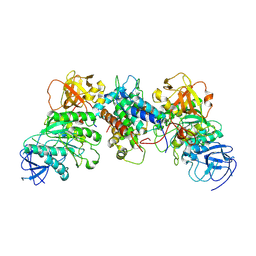 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 isoform 1, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
8J60
 
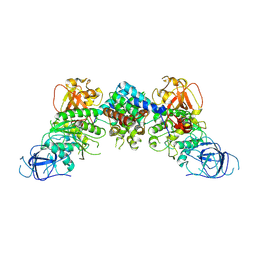 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 protein, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
3T9K
 
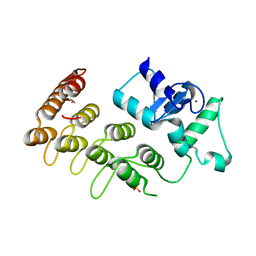 | | Crystal Structure of ACAP1 C-portion mutant S554D fused with integrin beta1 peptide | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1,Peptide from Integrin beta-1, SULFATE ION, ... | | Authors: | Sun, F, Pang, X, Zhang, K, Ma, J, Zhou, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
7DPA
 
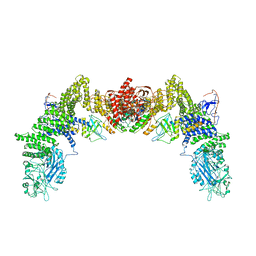 | | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Kaushik, R, Ehara, H, Yokoyama, T, Uchikubo-Kamo, T, Mishima-Tsumagari, C, Yonemochi, M, Ikeda, M, Hanada, K, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2020-12-18 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex.
Sci Adv, 7, 2021
|
|
5XG5
 
 | | Crystal structure of Mitsuba-1 with bound NAcGal | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MITSUBA-1 | | Authors: | Terada, D, Voet, A.R.D, Kamata, K, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Computational design of a symmetrical beta-trefoil lectin with cancer cell binding activity.
Sci Rep, 7, 2017
|
|
4OIY
 
 | | Crystal structure of Sec7p catalytic domain | | Descriptor: | MAGNESIUM ION, Protein transport protein SEC7 | | Authors: | Qiu, B, Zhang, K, Sun, F. | | Deposit date: | 2014-01-20 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | C-terminal motif within Sec7 domain regulates guanine nucleotide exchange activity via tuning protein conformation
Biochem.Biophys.Res.Commun., 446, 2014
|
|
7DVC
 
 | | Crystal structure of the computationally designed reDPBB_sym1 protein | | Descriptor: | ACETATE ION, CHLORIDE ION, reDPBB_sym1 protein | | Authors: | Yagi, S, Tagami, S, Padhi, A.K, Zhang, K.Y.J. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Seven Amino Acid Types Suffice to Create the Core Fold of RNA Polymerase.
J.Am.Chem.Soc., 143, 2021
|
|
7EZ2
 
 | | Holo L-16 ScaI Tetrahymena ribozyme | | Descriptor: | Holo L-16 ScaI Tetrahymena ribozyme, Holo L-16 ScaI Tetrahymena ribozyme S1, Holo L-16 ScaI Tetrahymena ribozyme S2, ... | | Authors: | Su, Z, Zhang, K, Kappel, K, Luo, B, Das, R, Chiu, W. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 angstrom resolution.
Nature, 596, 2021
|
|
7EZ0
 
 | | Apo L-21 ScaI Tetrahymena ribozyme | | Descriptor: | Apo L-21 ScaI Tetrahymena ribozyme, MAGNESIUM ION | | Authors: | Su, Z, Zhang, K, Kappel, K, Luo, B, Das, R, Chiu, W. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 angstrom resolution.
Nature, 596, 2021
|
|
7EVP
 
 | | Cryo-EM structure of the Gp168-beta-clamp complex | | Descriptor: | Beta sliding clamp, Sliding clamp inhibitor | | Authors: | Liu, B, Li, S, Liu, Y, Chen, H, Hu, Z, Wang, Z, Gou, L, Zhang, L, Ma, B, Wang, H, Matthews, S, Wang, Y, Zhang, K. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage Twort protein Gp168 is a beta-clamp inhibitor by occupying the DNA sliding channel.
Nucleic Acids Res., 49, 2021
|
|
7YCH
 
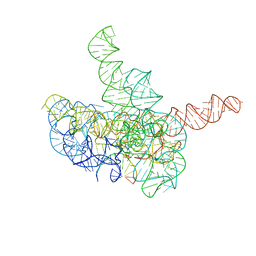 | | Cryo-EM structure of Tetrahymena ribozyme conformation 3 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (393-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7YCG
 
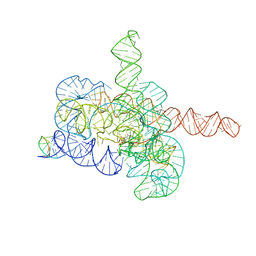 | | Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (393-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7YCI
 
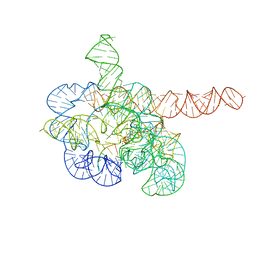 | | Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (389-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7YC8
 
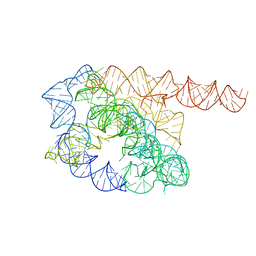 | | Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA (388-MER) | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7YG8
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 5 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (387-MER), RNA (5'-R(*CP*CP*CP*UP*C)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGD
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (384-MER), RNA (5'-R(*CP*C)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGC
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*UP*UP*AP*AP*CP*C)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7F02
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F04
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with Heme and ATP. | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F03
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
