8TW2
 
 | | Acinetobacter phage AP205 T=4 VLP | | Descriptor: | Coat protein | | Authors: | Meng, R, Xing, Z, Zhang, J. | | Deposit date: | 2023-08-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
8TOB
 
 | | Acinetobacter GP16 Type IV pilus | | Descriptor: | Fimbrial protein | | Authors: | Meng, R, Xing, Z, Zhang, J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
2L89
 
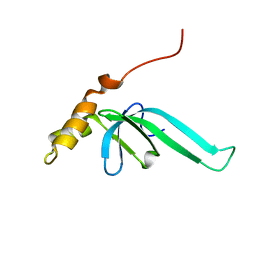 | |
5IZ8
 
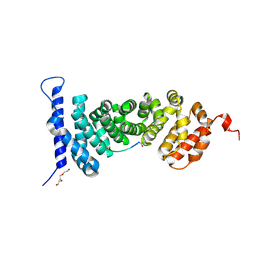 | | Protein-protein interaction | | Descriptor: | ACE-ALA-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein, TRIETHYLENE GLYCOL | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZA
 
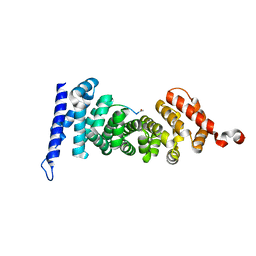 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-TRP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZ9
 
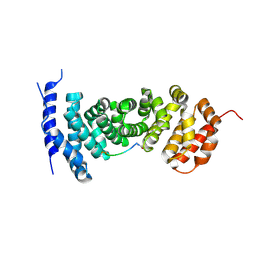 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
4EMP
 
 | | Crystal structure of the mutant of ClpP E137A from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Ye, F, Zhang, J, Liu, H, Luo, C, Yang, C.-G. | | Deposit date: | 2012-04-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Helix unfolding/refolding characterizes the functional dynamics of Staphylococcus aureus Clp protease
J.Biol.Chem., 288, 2013
|
|
2LNW
 
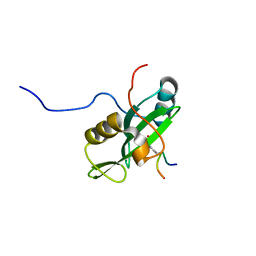 | | Identification and structural basis for a novel interaction between Vav2 and Arap3 | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3, Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
2LNX
 
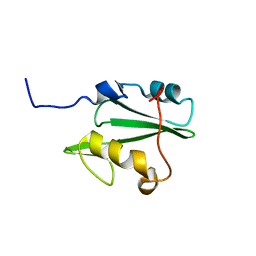 | | Solution structure of Vav2 SH2 domain | | Descriptor: | Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
2DK9
 
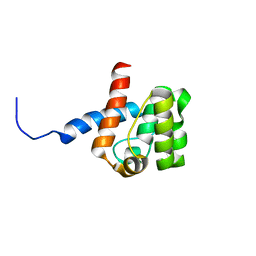 | | Solution structure of Calponin Homology domain of Human MICAL-1 | | Descriptor: | NEDD9-interacting protein with calponin homology and LIM domains | | Authors: | Sun, H, Dai, H, Zhang, J, Xiong, S, Wu, J, Shi, Y. | | Deposit date: | 2006-04-07 | | Release date: | 2006-09-19 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calponin homology domain of Human MICAL-1
J.Biomol.Nmr, 36, 2006
|
|
2AX5
 
 | | Solution Structure of Urm1 from Saccharomyces Cerevisiae | | Descriptor: | Hypothetical 11.0 kDa protein in FAA3-MAS3 intergenic region | | Authors: | Xu, J, Huang, H, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2005-09-03 | | Release date: | 2006-06-27 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Urm1 and its implications for the origin of protein modifiers.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4APP
 
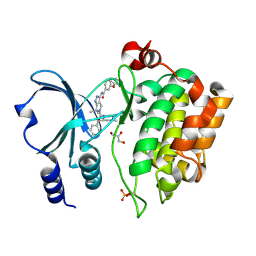 | | Crystal Structure of the Human p21-Activated Kinase 4 in Complex with (S)-N-(5-(3-benzyl-1-methylpiperazine-4-carbonyl)-6,6-dimethyl-1,4,5, 6-tetrahydropyrrolo(3,4-c)pyrazol-3-yl)-3-phenoxybenzamide | | Descriptor: | GLYCEROL, N-[6,6-dimethyl-5-[(2S)-4-methyl-2-(phenylmethyl)piperazin-1-yl]carbonyl-2,4-dihydropyrrolo[3,4-c]pyrazol-3-yl]-3-phenoxy-benzamide, SERINE/THREONINE-PROTEIN KINASE PAK 4 | | Authors: | Knighton, D.D, Deng, Y.L, Wang, C, Guo, C, McAlpine, I, Zhang, J, Kephart, S, Johnson, M.C, Li, H, Bouzida, D, Yang, A, Dong, L, Marakovits, J, Tikhe, J, Richardson, P, Guo, L.C, Kania, R, Edwards, M.P, Kraynov, E, Christensen, J, Piraino, J, Lee, J, Dagostino, E, Del-Carmen, C, Smeal, T, Murray, B.W. | | Deposit date: | 2012-04-04 | | Release date: | 2012-06-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Pyrroloaminopyrazoles as Novel Pak Inhibitors.
J.Med.Chem., 55, 2012
|
|
5JIQ
 
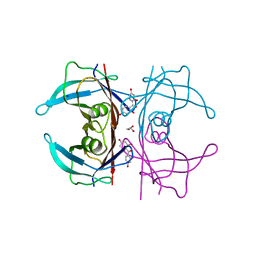 | | Crystal Structure of Human Transthyretin in Complex with 2,2',4,4'-tetrahydroxybenzophenone (BP2) | | Descriptor: | GLYCEROL, SODIUM ION, Transthyretin, ... | | Authors: | Begum, A, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | Deposit date: | 2016-04-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
5JIM
 
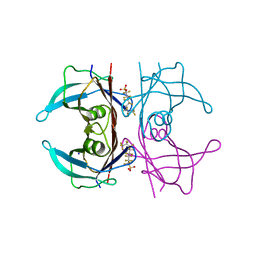 | | Crystal Structure of Human Transthyretin in Complex with Perfluoroktansulfonsyra (PFOS) | | Descriptor: | SODIUM ION, Transthyretin, heptadecafluoro-1-octanesulfonic acid | | Authors: | Begum, A, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | Deposit date: | 2016-04-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
7TQA
 
 | | Crystal Structure of monoclonal S9.6 Fab | | Descriptor: | Fab S9.6 heavy chain, Fab S9.6 light chain, GLYCEROL, ... | | Authors: | Bou-Nader, C, Zhang, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Structural basis of R-loop recognition by the S9.6 monoclonal antibody.
Nat Commun, 13, 2022
|
|
7TQB
 
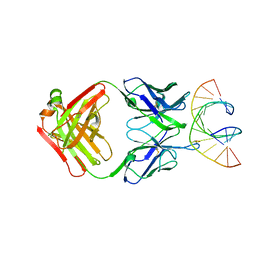 | |
1YOP
 
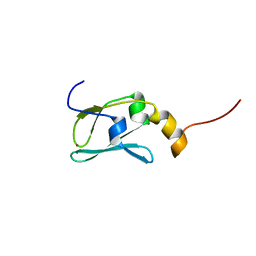 | | The solution structure of Kti11p | | Descriptor: | Kti11p, ZINC ION | | Authors: | Sun, J, Zhang, J, Wu, F, Xu, C, Li, S, Zhao, W, Wu, Z, Wu, J, Zhou, C.-Z, Shi, Y. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module.
Biochemistry, 44, 2005
|
|
6JJP
 
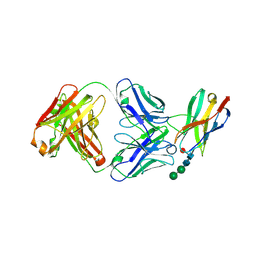 | | Crystal structure of Fab of a PD-1 monoclonal antibody MW11-h317 in complex with PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of MW11-h317, Programmed cell death protein 1, ... | | Authors: | Wang, M, Wang, J, Wang, R, Jiao, S, Wang, S, Zhang, J, Zhang, M. | | Deposit date: | 2019-02-26 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of a monoclonal antibody that targets PD-1 in a manner requiring PD-1 Asn58 glycosylation.
Commun Biol, 2, 2019
|
|
3MGM
 
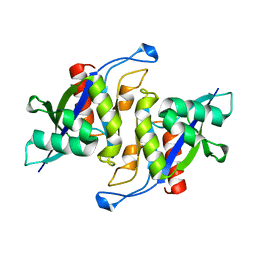 | | Crystal structure of human NUDT16 | | Descriptor: | U8 snoRNA-decapping enzyme | | Authors: | Yan, J, Lu, G, Zhang, J, Qi, J, Li, Z, Gao, F. | | Deposit date: | 2010-04-07 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure and the template mRNA decapping activity of human NUDT16
To be Published
|
|
3IZ3
 
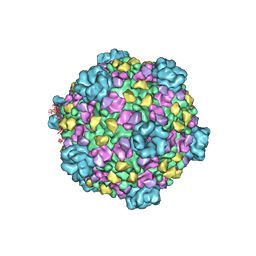 | | CryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Structural protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Cheng, L, Sun, J, Zhang, K, Mou, Z, Huang, X, Ji, G, Sun, F, Zhang, J, Zhu, P. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3J32
 
 | | An asymmetric unit map from electron cryo-microscopy of Haliotis diversicolor molluscan hemocyanin isoform 1 (HdH1) | | Descriptor: | Hemocyanin isoform 1 | | Authors: | Zhang, Q, Dai, X, Cong, Y, Zhang, J, Chen, D.-H, Dougherty, M, Wang, J, Ludtke, S, Schmid, M.F, Chiu, W. | | Deposit date: | 2013-02-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of a molluscan hemocyanin suggests its allosteric mechanism.
Structure, 21, 2013
|
|
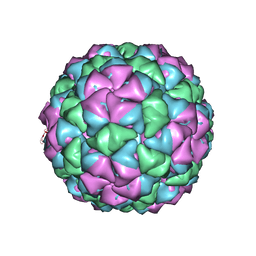
5KIP
 
 | |
8KD3
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KC7
 
 | | Rpd3S histone deacetylase complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-06 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD2
 
 | | Rpd3S in complex with 187bp nucleosome | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
