7XOV
 
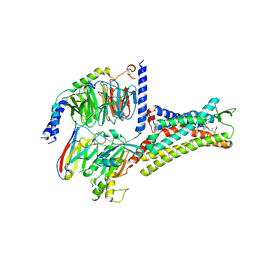 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | 2-[2-[[4-(4-chloranyl-2,5-dimethoxy-phenyl)-5-(2-cyclohexylethyl)-1,3-thiazol-2-yl]carbamoyl]-5,7-dimethyl-indol-1-yl]ethanoic acid, Cholecystokinin receptor type A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
4O5P
 
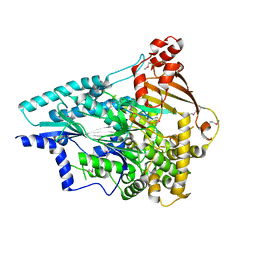 | | Crystal structure of an uncharacterized protein from Pseudomonas aeruginosa | | Descriptor: | Uncharacterized protein | | Authors: | Hu, H.D, Gao, Z.Q, Zhang, H, Dong, Y.H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structure of the type VI secretion phospholipase effector Tle1 provides insight into its hydrolysis and membrane targeting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4F3L
 
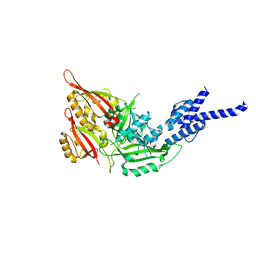 | | Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex | | Descriptor: | BMAL1b, Circadian locomoter output cycles protein kaput | | Authors: | Huang, N, Chelliah, Y, Shan, Y, Taylor, C, Yoo, S, Partch, C, Green, C.B, Zhang, H, Takahashi, J. | | Deposit date: | 2012-05-09 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex.
Science, 337, 2012
|
|
6NYG
 
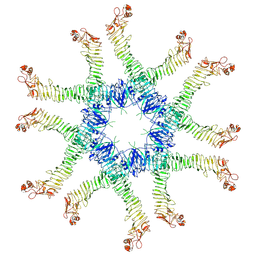 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2a (OA-2a) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYF
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 1 (OA-1) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYM
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2d (OA-2d) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8JYU
 
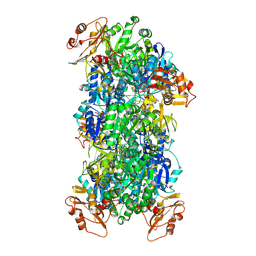 | | Acyl-ACP Synthetase structure bound to Decanoyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, DECANOIC ACID, ... | | Authors: | Huang, H, Chang, S, Huang, M, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Acyl-ACP Synthetase structure bound to Decanoyl-AMP
To Be Published
|
|
8JYL
 
 | | Acyl-ACP Synthetase structure bound to C10-AMS | | Descriptor: | Acyl-acyl carrier protein synthetase, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-decanoylsulfamate | | Authors: | Huang, H, Chang, S, Huang, M, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Acyl-ACP Synthetase structure bound to C10-AMS
To Be Published
|
|
6NYJ
 
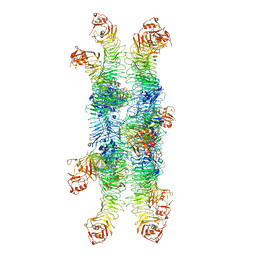 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2b (OA-2b) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYN
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2e (OA-2e) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7BPB
 
 | | Human AAA+ ATPase VCP mutant - T76E, AMP-PNP bound form, Conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-22 | | Release date: | 2021-03-31 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7BP9
 
 | | Human AAA+ ATPase VCP mutant - T76E, ADP-bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7BPA
 
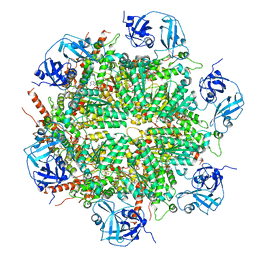 | | Human AAA+ ATPase VCP mutant - T76A, AMP-PNP-bound form, Conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7BP8
 
 | | Human AAA+ ATPase VCP mutant - T76A, ADP-bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
4HE9
 
 | | Crystal Structure of HIV-1 protease mutants I54M complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 protease, ... | | Authors: | Shen, C.H, Zhang, H, Weber, I.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
4HDP
 
 | | Crystal Structure of HIV-1 protease mutants I50V complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 Protease, ... | | Authors: | Shen, C.H, Zhang, H, Weber, I.T. | | Deposit date: | 2012-10-02 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
6NYL
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2c (OA-2c) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8GU6
 
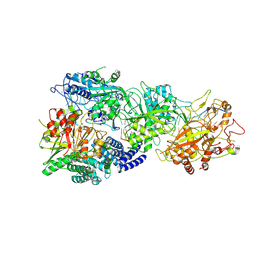 | | Structure of the SbCas7-11-crRNA-NTR-Csx29 complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-09-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Target RNA-guided protease activity in type III-E CRISPR-Cas system.
Nucleic Acids Res., 50, 2022
|
|
8GNA
 
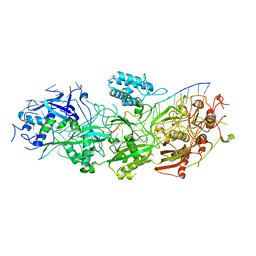 | | Structure of the SbCas7-11-crRNA-NTR complex | | Descriptor: | RAMP superfamily protein, RNA (32-MER), RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*U)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Target RNA-guided protease activity in type III-E CRISPR-Cas system.
Nucleic Acids Res., 50, 2022
|
|
4OQ6
 
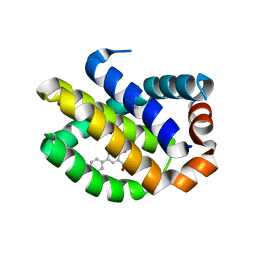 | | Crystal Structure of Human MCL-1 Bound to Inhibitor 4-hydroxy-4'-propylbiphenyl-3-carboxylic acid | | Descriptor: | 4-hydroxy-4'-propylbiphenyl-3-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Petros, A.M, Swann, S.L, Song, D, Swinger, K, Park, C, Zhang, H, Wendt, M.D, Kunzer, A.R, Souers, A.J, Sun, C. | | Deposit date: | 2014-02-07 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-based discovery of potent inhibitors of the anti-apoptotic MCL-1 protein.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6ITW
 
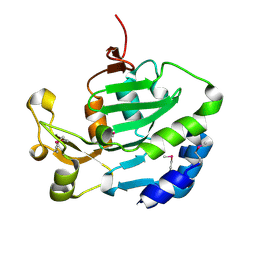 | |
4HZ9
 
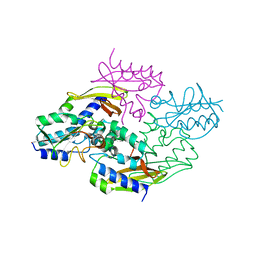 | | Crystal structure of the type VI native effector-immunity complex Tae3-Tai3 from Ralstonia pickettii | | Descriptor: | Putative cytoplasmic protein, Putative periplasmic protein | | Authors: | Dong, C, Zhang, H, Gao, Z.Q, Dong, Y.H. | | Deposit date: | 2012-11-14 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the inhibition of type VI effector Tae3 by its immunity protein Tai3
Biochem.J., 454, 2013
|
|
4HZB
 
 | | Crystal structure of the type VI SeMet effector-immunity complex Tae3-Tai3 from Ralstonia pickettii | | Descriptor: | Putative cytoplasmic protein, Putative periplasmic protein | | Authors: | Dong, C, Zhang, H, Gao, Z.Q, Dong, Y.H. | | Deposit date: | 2012-11-15 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the inhibition of type VI effector Tae3 by its immunity protein Tai3
Biochem.J., 454, 2013
|
|
7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MYN
 
 | | Cryo-EM Structure of p110alpha in complex with p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
