5JPP
 
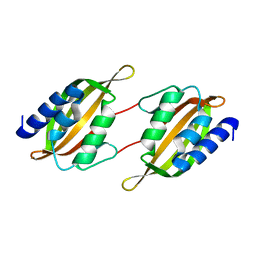 | | Structure of limonene epoxide hydrolase mutant - H-2-H5 | | Descriptor: | limonene epoxide hydrolase | | Authors: | Li, G, Zhang, H, Sun, Z, Liu, X, Reetz, M.T. | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multi-Parameter Optimization in Directed Evolution: Engineering Thermostability, Enantioselectivity and Activity of an Epoxide Hydrolase
To be published
|
|
1IJL
 
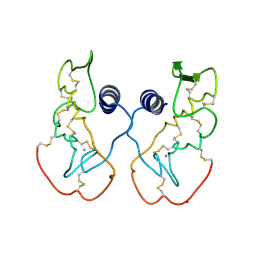 | | Crystal structure of acidic phospholipase A2 from deinagkistrodon acutus | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2, ZINC ION | | Authors: | Gu, L, Zhang, H, Song, S, Zhou, Y, Lin, Z. | | Deposit date: | 2001-04-27 | | Release date: | 2001-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an acidic phospholipase A2 from the venom of Deinagkistrodon acutus.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4I5X
 
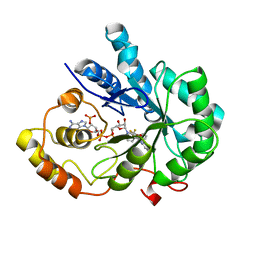 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Flufenamic acid | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Zhang, H, Zhao, Y. | | Deposit date: | 2012-11-29 | | Release date: | 2013-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
1M4M
 
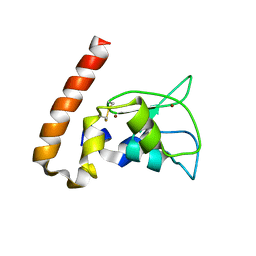 | | Mouse Survivin | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, ZINC ION | | Authors: | Muchmore, S.W, Chen, J, Jakob, C, Zakula, D, Matayoshi, E.D, Wu, W, Zhang, H, Li, F, Ng, S.C, Altieri, D.C. | | Deposit date: | 2002-07-03 | | Release date: | 2002-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE AND MUTAGENIC ANALYSIS OF THE INHIBITOR-OF-APOPTOSIS PROTEIN SURVIVIN
MOL.CELL, 6, 2000
|
|
1P4M
 
 | | CRYSTAL STRUCTURE OF RIBOFLAVIN KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Karthikeyan, S, Zhou, Q, Mseeh, F, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-04-23 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Riboflavin Kinase Reveals a Beta Barrel Fold and a Novel Active Site Arch
Structure, 11, 2003
|
|
1FWL
 
 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE | | Descriptor: | HOMOSERINE KINASE | | Authors: | Zhou, T, Daugherty, M, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2000-09-22 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and mechanism of homoserine kinase: prototype for the GHMP kinase superfamily.
Structure Fold.Des., 8, 2000
|
|
1FWK
 
 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE COMPLEXED WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HOMOSERINE KINASE, MAGNESIUM ION | | Authors: | Zhou, T, Daugherty, M, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2000-09-22 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of homoserine kinase: prototype for the GHMP kinase superfamily.
Structure Fold.Des., 8, 2000
|
|
1NB9
 
 | | Crystal Structure of Riboflavin Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RIBOFLAVIN, ... | | Authors: | Karthikeyan, S, Zhou, Q, Mseeh, F, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Riboflavin Kinase Reveals a Beta Barrel Fold and a Novel Active Site Arch
Structure, 11, 2003
|
|
1M32
 
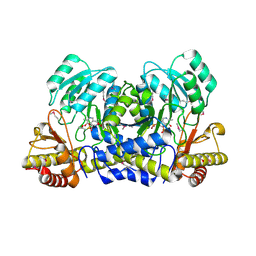 | | Crystal Structure of 2-aminoethylphosphonate Transaminase | | Descriptor: | 2-aminoethylphosphonate-pyruvate aminotransferase, PHOSPHATE ION, PHOSPHONOACETALDEHYDE, ... | | Authors: | Chen, C.C.H, Zhang, H, Kim, A.D, Howard, A, Sheldrick, G.M, Mariano-Dunnaway, D, Herzberg, O. | | Deposit date: | 2002-06-26 | | Release date: | 2002-11-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Degradation Pathway of the Phosphonate Ciliatine: Crystal Structure of 2-Aminoethylphosphonate Transaminase
Biochemistry, 41, 2002
|
|
1EET
 
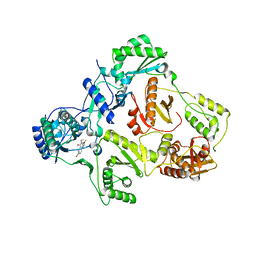 | | HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH THE INHIBITOR MSC204 | | Descriptor: | 1-(5-BROMO-PYRIDIN-2-YL)-3-[2-(6-FLUORO-2-HYDROXY-3-PROPIONYL-PHENYL)-CYCLOPROPYL]-UREA, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Hogberg, M, Sahlberg, C, Engelhardt, P, Noreen, R, Kangasmetsa, J, Johansson, N.G, Oberg, B, Vrang, L, Zhang, H, Sahlberg, B.L, Unge, T, Lovgren, S, Fridborg, K, Backbro, K. | | Deposit date: | 2000-02-03 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Urea-PETT compounds as a new class of HIV-1 reverse transcriptase inhibitors. 3. Synthesis and further structure-activity relationship studies of PETT analogues.
J.Med.Chem., 42, 1999
|
|
1MXI
 
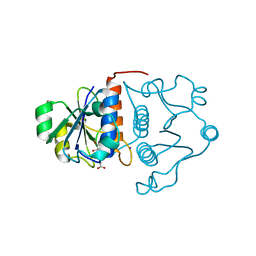 | | Structure of YibK from Haemophilus influenzae (HI0766): a Methyltransferase with a Cofactor Bound at a Site Formed by a Knot | | Descriptor: | Hypothetical tRNA/rRNA methyltransferase HI0766, IODIDE ION, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-10-02 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): a Cofactor Bound at a Site Formed by a Knot
Proteins, 51, 2003
|
|
1G5J
 
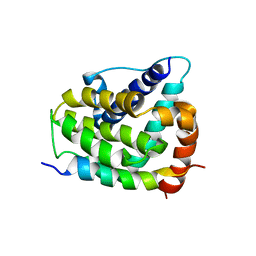 | | COMPLEX OF BCL-XL WITH PEPTIDE FROM BAD | | Descriptor: | APOPTOSIS REGULATOR BCL-X, BAD PROTEIN | | Authors: | Petros, A.M, Nettesheim, D.G, Wang, Y, Olejniczak, E.T, Meadows, R.P, Mack, J, Swift, K, Matayoshi, E.D, Zhang, H, Thompson, C.B, Fesik, S.W. | | Deposit date: | 2000-11-01 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies.
Protein Sci., 9, 2000
|
|
1NB0
 
 | | Crystal Structure of Human Riboflavin Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, hypothetical protein FLJ11149 | | Authors: | Karthikeyan, S, Zhou, Q, Mseeh, F, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-12-01 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Riboflavin Kinase Reveals a Beta Barrel Fold and a Novel Active Site Arch
Structure, 11, 2003
|
|
1NI1
 
 | | Imidazole and cyanophenyl farnesyl transferase inhibitors | | Descriptor: | 2-CHLORO-5-(3-CHLORO-PHENYL)-6-[(4-CYANO-PHENYL)-(3-METHYL-3H-IMIDAZOL-4-YL)- METHOXYMETHYL]-NICOTINONITRILE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Tong, Y, Lin, N.H, Wang, L, Hasvold, L, Wang, W, Leonard, N, Li, T, Li, Q, Cohen, J, Gu, W.Z, Zhang, H, Stoll, V, Bauch, J, Marsh, K, Rosenberg, S.H, Sham, H.L. | | Deposit date: | 2002-12-20 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of potent imidazole and cyanophenyl containing farnesyltransferase inhibitors with improved oral bioavailability.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1GQ2
 
 | | Malic Enzyme from Pigeon Liver | | Descriptor: | CHLORIDE ION, MALIC ENZYME, MANGANESE (II) ION, ... | | Authors: | Yang, Z, Zhang, H, Liang, T. | | Deposit date: | 2001-11-19 | | Release date: | 2002-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of the Pigeon Cytosolic Nadp+ -Dependent Malic Enzyme
Protein Sci., 11, 2002
|
|
1Q9S
 
 | | Crystal structure of riboflavin kinase with ternary product complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Karthikeyan, S, Zhou, Q, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-08-25 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Ligand binding-induced conformational changes in riboflavin kinase: structural basis for the ordered mechanism.
Biochemistry, 42, 2003
|
|
4XWT
 
 | | Crystal structure of RNase J complexed with UMP | | Descriptor: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | Deposit date: | 2015-01-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
5Z2S
 
 | | Crystal structure of DUX4-HD2 domain | | Descriptor: | Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
5Z2T
 
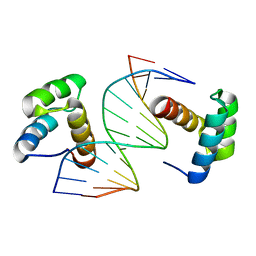 | | Crystal structure of DNA-bound DUX4-HD2 | | Descriptor: | 5'-D(*TP*TP*CP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*TP*T)-3', 5'-D(P*AP*AP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*GP*T)-3', Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-04 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
1IM8
 
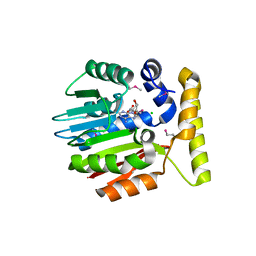 | | Crystal structure of YecO from Haemophilus influenzae (HI0319), a methyltransferase with a bound S-adenosylhomocysteine | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOSELENOCYSTEINE, YecO | | Authors: | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-10 | | Release date: | 2001-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of YecO from Haemophilus influenzae (HI0319) reveals a methyltransferase fold and a bound S-adenosylhomocysteine.
Proteins, 45, 2001
|
|
1J85
 
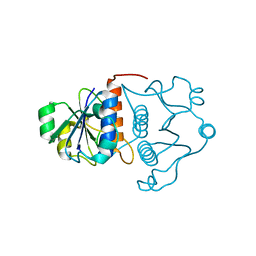 | | Structure of YibK from Haemophilus influenzae (HI0766), a truncated sequence homolog of tRNA (guanosine-2'-O-) methyltransferase (SpoU) | | Descriptor: | YibK | | Authors: | Lim, K, Zhang, H, Toedt, J, Tempcyzk, A, Krajewski, W, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-20 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae
(HI0766): A cofactor bound at a site formed by a knot
Proteins, 51, 2003
|
|
1KQN
 
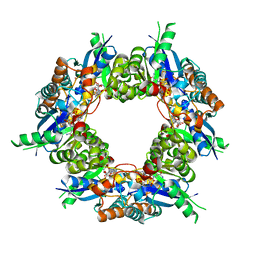 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XENON | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Human Nicotinamide/Nicotonic Acid Mononucleotide Adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2003
|
|
1KR2
 
 | | CRYSTAL STRUCTURE OF HUMAN NMN/NAMN ADENYLYL TRANSFERASE COMPLEXED WITH TIAZOFURIN ADENINE DINUCLEOTIDE (TAD) | | Descriptor: | BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
1KQO
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with deamido-NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
1W5V
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DI-3-FLUORO-BENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
