5HNU
 
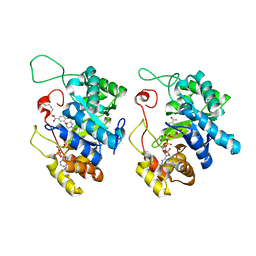 | | Crystal Structure of AKR1C3 complexed with octyl gallate | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, octyl 3,4,5-trihydroxybenzoate | | Authors: | Li, C, Zhao, Y, Zhang, H, Hu, X. | | Deposit date: | 2016-01-18 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of AKR1C3 complexed with octyl gallte
To Be Published
|
|
2MPX
 
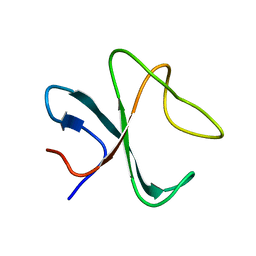 | |
5HNT
 
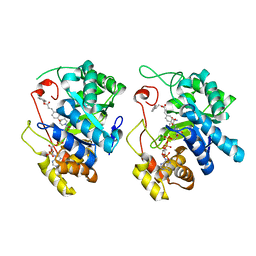 | | Crystal Structure of AKR1C3 complexed with CAPE | | Descriptor: | 2-phenylethyl (2E)-3-(3,4-dihydroxyphenyl)prop-2-enoate, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C, Zhao, Y, Zhang, H, Hu, X. | | Deposit date: | 2016-01-18 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of AKR1C3 complexed with octyl gallate
To Be Published
|
|
5IC0
 
 | | Structural analysis of a talin triple domain module | | Descriptor: | 1,2-ETHANEDIOL, Talin-1 | | Authors: | Wu, J, Chang, Y.-C.E, Zhang, H, Brennan, M.L. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Functional Analysis of a Talin Triple-Domain Module Suggests an Alternative Talin Autoinhibitory Configuration.
Structure, 24, 2016
|
|
5FB7
 
 | |
5IC1
 
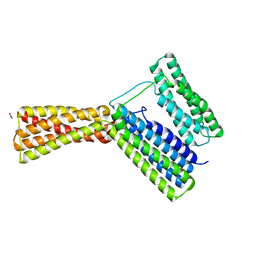 | | Structural analysis of a talin triple domain module, E1794Y, E1797Y, Q1801Y mutant | | Descriptor: | 1,2-ETHANEDIOL, Talin-1 | | Authors: | Wu, J, Chang, Y.-C.E, Zhang, H, Huang, Q.-Q. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of a Talin Triple-Domain Module Suggests an Alternative Talin Autoinhibitory Configuration.
Structure, 24, 2016
|
|
5H48
 
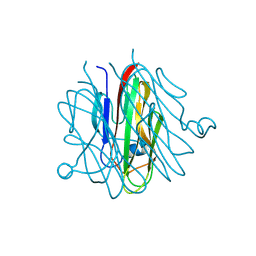 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H4C
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein Cbln4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5I8J
 
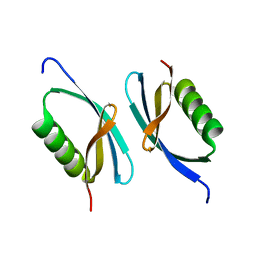 | | Crystal structure of Dmd from phage RB69 | | Descriptor: | Dmd discriminator of mRNA degradation | | Authors: | Wei, Y, Zhang, H, Gao, Z.Q, Dong, Y.H. | | Deposit date: | 2016-02-19 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterizations of phage antitoxin Dmd and its interactions with bacterial toxin RnlA
Biochem. Biophys. Res. Commun., 472, 2016
|
|
2MWN
 
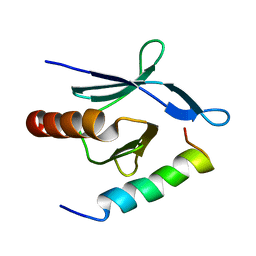 | | Talin-F3 / RIAM N-terminal Peptide complex | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein, Talin-1 | | Authors: | Yang, J, Zhu, L, Zhang, H, Hirbawi, J, Fukuda, K, Dwivedi, P, Liu, J, Byzova, T, Plow, E.F, Wu, J, Qin, J. | | Deposit date: | 2014-11-13 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational activation of talin by RIAM triggers integrin-mediated cell adhesion.
Nat Commun, 5, 2014
|
|
7WIG
 
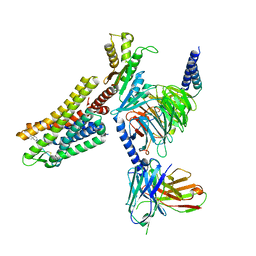 | | Cryo-EM structure of the L-054,264-bound human SSTR2-Gi1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Chen, L, Wang, W, Dong, Y, Shen, D, Guo, J, Qin, J, Zhang, H, Shen, Q, Zhang, Y, Mao, C. | | Deposit date: | 2022-01-03 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of the endogenous peptide- and selective non-peptide agonist-bound SSTR2 signaling complexes.
Cell Res., 32, 2022
|
|
7WIC
 
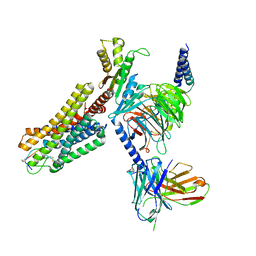 | | Cryo-EM structure of the SS-14-bound human SSTR2-Gi1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Chen, L, Wang, W, Dong, Y, Shen, D, Guo, J, Qin, J, Zhang, H, Shen, Q, Zhang, Y, Mao, C. | | Deposit date: | 2022-01-03 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of the endogenous peptide- and selective non-peptide agonist-bound SSTR2 signaling complexes.
Cell Res., 32, 2022
|
|
8HEF
 
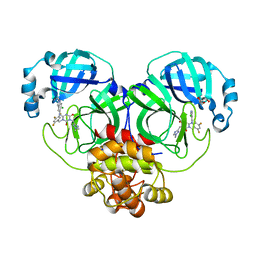 | | The Crystal structure of deuterated S-217622 (Ensitrelvir) bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Yan, M, Zhang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Synthesis of deuterated S-217622 (Ensitrelvir) with antiviral activity against coronaviruses including SARS-CoV-2.
Antiviral Res., 213, 2023
|
|
8W9X
 
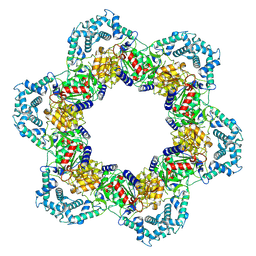 | |
8W9L
 
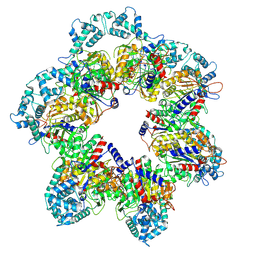 | |
8WIV
 
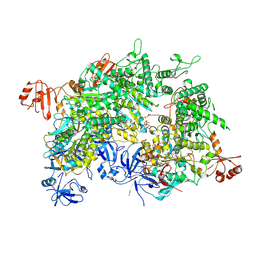 | | Cryo-EM structure of a bacterial protein | | Descriptor: | Helicase HerA central domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WJ3
 
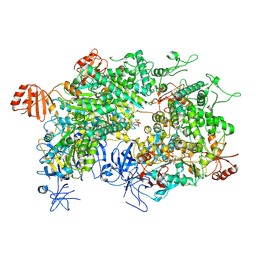 | | Cryo-EM structure of a bacterial protein | | Descriptor: | Helicase HerA central domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WLD
 
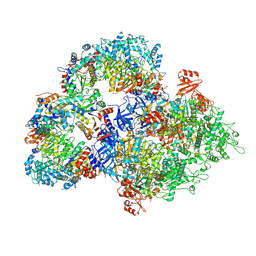 | | Cryo-EM structure of SIR2/HerA antiphage complex | | Descriptor: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-09-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Cryo-EM structure of SIR2/HerA antiphage complex
To Be Published
|
|
8WOC
 
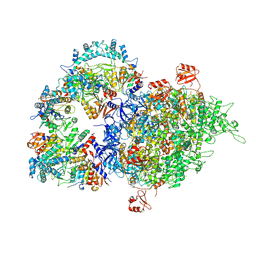 | | Cryo-EM structure of SIR2/HerA complex | | Descriptor: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WOD
 
 | | Cryo-EM structure of SIR2/HerA complex | | Descriptor: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WK0
 
 | | Cryo-EM structure of a bacterial protein | | Descriptor: | Helicase HerA central domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-09-26 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WOF
 
 | | Cryo-EM structure of SIR2/HerA complex | | Descriptor: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
7CZ5
 
 | | Cryo-EM structure of the human growth hormone-releasing hormone receptor-Gs protein complex | | Descriptor: | CHOLESTEROL, Growth hormone-releasing hormone receptor,growth hormone-releasing hormone receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhou, F, Zhang, H, Cong, Z, Zhao, L, Zhou, Q, Mao, C, Cheng, X, Shen, D, Cai, X, Ma, C, Wang, Y, Dai, A, Zhou, Y, Sun, W, Zhao, F, Zhao, S, Jiang, H, Jiang, Y, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-07 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for activation of the growth hormone-releasing hormone receptor.
Nat Commun, 11, 2020
|
|
7W3Z
 
 | | Cryo-EM Structure of Human Gastrin Releasing Peptide Receptor in complex with the agonist Gastrin Releasing Peptide and Gq heterotrimers | | Descriptor: | Gastrin Releasing Peptide PRGNHWAVGHLM(NH2), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhan, Y, Peng, S, Zhang, H. | | Deposit date: | 2021-11-26 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of human gastrin-releasing peptide receptors bound to antagonist and agonist for cancer and itch therapy.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7W40
 
 | | Cryo-EM Structure of Human Gastrin Releasing Peptide Receptor in complex with the agonist Bombesin (6-14) [D-Phe6, beta-Ala11, Phe13, Nle14] and Gq heterotrimers | | Descriptor: | Bombesin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhan, Y, Peng, S, Zhang, H. | | Deposit date: | 2021-11-26 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of human gastrin-releasing peptide receptors bound to antagonist and agonist for cancer and itch therapy.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
