3TNB
 
 | | Crystal structure of GkaP mutant G209D/R230H from Geobacillus kaustophilus HTA426 | | 分子名称: | COBALT (II) ION, Phosphotriesterase | | 著者 | An, J, Zhang, Z, Zhang, Y, Feng, Y, Wu, G. | | 登録日 | 2011-09-01 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Engineering a thermostable lactonase for enhanced phosphotriesterase activity against organophosphate pesticides
to be published
|
|
8T9O
 
 | |
8T9P
 
 | |
8T9Q
 
 | |
6J76
 
 | | Structure of 3,6-anhydro-L-galactose Dehydrogenase in Complex with NAP | | 分子名称: | Aldehyde dehydrogenase A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, P.Y, Wang, Y, Chen, X.L, Zhang, Y.Z. | | 登録日 | 2019-01-17 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.368 Å) | | 主引用文献 | 3,6-Anhydro-L-Galactose Dehydrogenase VvAHGD is a Member of a New Aldehyde Dehydrogenase Family and Catalyzes by a Novel Mechanism with Conformational Switch of Two Catalytic Residues Cysteine 282 and Glutamate 248.
J.Mol.Biol., 432, 2020
|
|
7CAL
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Inwardly Rectifying Potassium Channel KAT1 from Arabidopsis | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Potassium channel KAT1 | | 著者 | Li, S.Y, Yang, F, Sun, D.M, Zhang, Y, Zhang, M.G, Zhou, P, Liu, S.L, Zhang, Y.N, Zhang, L.H, Tian, C.L. | | 登録日 | 2020-06-09 | | 公開日 | 2020-07-29 | | 最終更新日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of the hyperpolarization-activated inwardly rectifying potassium channel KAT1 from Arabidopsis.
Cell Res., 30, 2020
|
|
7WDL
 
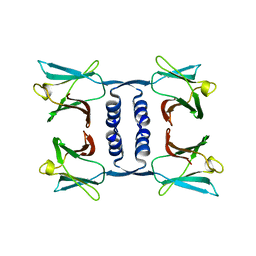 | | Fungal immunomodulatory protein FIP-nha | | 分子名称: | Fungal immunomodulatory protein | | 著者 | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.903 Å) | | 主引用文献 | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties.
Int.J.Biol.Macromol., 213, 2022
|
|
7CQQ
 
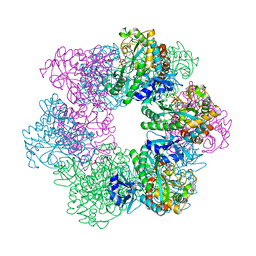 | | GmaS in complex with AMPPNP and MetSox | | 分子名称: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, C.Y, Zhang, Y.Z. | | 登録日 | 2020-08-11 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.295 Å) | | 主引用文献 | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
4F6V
 
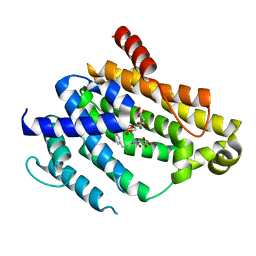 | | Crystal structure of dehydrosqualene synthase (crtm) from s. aureus complexed with bph-1034, mg2+ and fmp. | | 分子名称: | (2E,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl dihydrogen phosphate, 2,4-dioxo-4-{[3-(3-phenoxyphenyl)propyl]amino}butanoic acid, Dehydrosqualene synthase, ... | | 著者 | Lin, F.-Y, Zhang, Y, Liu, Y.-L, Oldfield, E. | | 登録日 | 2012-05-15 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
7CQL
 
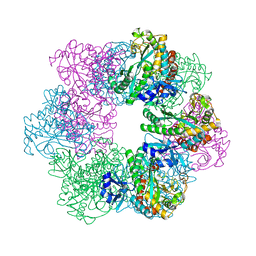 | | Apo GmaS without ligand | | 分子名称: | Type III glutamate--ammonia ligase | | 著者 | Li, C.Y, Zhang, Y.Z. | | 登録日 | 2020-08-11 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
4F6X
 
 | | Crystal structure of dehydrosqualene synthase (crtm) from s. aureus complexed with bph-1112 | | 分子名称: | 1-[3-(hexyloxy)benzyl]-4-hydroxy-2-oxo-1,2-dihydropyridine-3-carboxylic acid, D(-)-TARTARIC ACID, Dehydrosqualene synthase, ... | | 著者 | Lin, F.-Y, Zhang, Y, Liu, Y.-L, Oldfield, E. | | 登録日 | 2012-05-15 | | 公開日 | 2012-06-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
7CQN
 
 | | GmaS in complex with AMPPCP | | 分子名称: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Type III glutamate--ammonia ligase | | 著者 | Li, C.Y, Zhang, Y.Z. | | 登録日 | 2020-08-11 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.962 Å) | | 主引用文献 | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7C8J
 
 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | 分子名称: | Angiotensin-converting enzyme, SARS-CoV-2 Receptor binding domain, ZINC ION | | 著者 | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | 登録日 | 2020-06-01 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7WJS
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13157 | | 分子名称: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-07 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WL4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor SLP-50 | | 分子名称: | Bromodomain-containing protein 4, ~{N}-[2-ethyl-6-(4-methylpiperazin-1-yl)-3-oxidanylidene-2,7-diazatricyclo[6.3.1.0^{4,12}]dodeca-1(12),4,6,8,10-pentaen-9-yl]-2,4-bis(fluoranyl)benzenesulfonamide | | 著者 | Zhang, C, Wang, C, Li, W, Zhang, Y, Xu, Y, Sun, L. | | 登録日 | 2022-01-12 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Design, synthesis, and anticancer evaluation of ammosamide B with pyrroloquinoline derivatives as novel BRD4 inhibitors.
Bioorg.Chem., 127, 2022
|
|
7WNI
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13158 | | 分子名称: | 1,2-ETHANEDIOL, 7-[2-[2,4-bis(fluoranyl)phenoxy]-5-(2-oxidanylpropan-2-yl)phenyl]-2-[4-(2-hydroxyethyloxy)-3,5-dimethyl-phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-18 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.12 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WKY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13153 | | 分子名称: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-12 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WMU
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13146 | | 分子名称: | 1,2-ETHANEDIOL, Isoform 4 of Bromodomain-containing protein 2, ~{N}-[4-[2,4-bis(fluoranyl)phenoxy]-3-[2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-5-methyl-4-oxidanylidene-furo[3,2-c]pyridin-7-yl]phenyl]ethanesulfonamide | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-17 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WN5
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13142 | | 分子名称: | 1,2-ETHANEDIOL, Isoform 4 of Bromodomain-containing protein 2, POTASSIUM ION, ... | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-17 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WLN
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13153 | | 分子名称: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-13 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WNA
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13120 | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Isoform 4 of Bromodomain-containing protein 2, ... | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-17 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WMQ
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13157 | | 分子名称: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, FORMIC ACID, GLYCEROL, ... | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-16 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7CQU
 
 | | GmaS/ADP/MetSox-P complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MAGNESIUM ION, ... | | 著者 | Li, C.Y, Zhang, Y.Z. | | 登録日 | 2020-08-11 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
5V1X
 
 | | Carbon Sulfoxide lyase, Egt2 Y134F in complex with its substrate | | 分子名称: | (1S)-2-{2-[(R)-(2R)-2-amino-2-carboxyethanesulfinyl]-1H-imidazol-4-yl}-1-carboxy-N,N,N-trimethylethan-1-aminium, FORMIC ACID, Hercynylcysteine sulfoxide lyase | | 著者 | Irani, S, Zhang, Y. | | 登録日 | 2017-03-02 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.558 Å) | | 主引用文献 | Snapshots of C-S Cleavage in Egt2 Reveals Substrate Specificity and Reaction Mechanism.
Cell Chem Biol, 25, 2018
|
|
7CQX
 
 | | GmaS/ADP complex-Conformation 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | 著者 | Li, C.Y, Zhang, Y.Z. | | 登録日 | 2020-08-11 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
