7YCI
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (389-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7YCG
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (393-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7YCH
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 3 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (393-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7V66
 
 | | Structure of Apoferritin | | Descriptor: | Ferritin heavy chain | | Authors: | Zhang, X, Wu, C, Shi, H. | | Deposit date: | 2021-08-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (1.89 Å) | | Cite: | Low-cooling-rate freezing in biomolecular cryo-electron microscopy for recovery of initial frames.
QRB Discov, 2, 2021
|
|
7V4V
 
 | |
7VH2
 
 | |
7VGQ
 
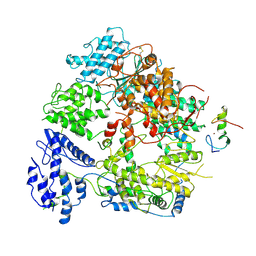 | | Cryo-EM structure of Machupo virus polymerase L in complex with matrix protein Z | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,RING finger protein Z, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Zhang, X, Ma, J, Zhang, S. | | Deposit date: | 2021-09-18 | | Release date: | 2021-09-29 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of Machupo virus polymerase in complex with matrix protein Z.
Nat Commun, 12, 2021
|
|
7VH1
 
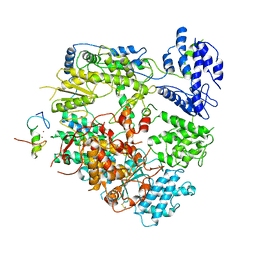 | | Cryo-EM structure of Machupo virus dimeric L-Z complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,RING finger protein Z, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Zhang, X, Ma, J, Zhang, S. | | Deposit date: | 2021-09-20 | | Release date: | 2021-09-29 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Machupo virus polymerase in complex with matrix protein Z.
Nat Commun, 12, 2021
|
|
7FFO
 
 | | Cryo-EM structure of VEEV VLP at the 5-fold axes | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFQ
 
 | | Cryo-EM structure of VEEV VLP at the 2-fold axes | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFL
 
 | | Cryo-EM structure of VEEV VLP-LDLRAD3-D1 complex at the 2-fold axes | | Descriptor: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFE
 
 | | Cryo-EM structure of VEEV VLP | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFN
 
 | | Cryo-EM structure of VEEV VLP-LDLRAD3-D1 complex at the 5-fold axes | | Descriptor: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7XGD
 
 | | Cryo-EM structure of Apo-IGF1R map 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin-like growth factor 1 receptor | | Authors: | Zhang, X, Wu, C. | | Deposit date: | 2022-04-04 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of Apo-IGF1R
To Be Published
|
|
5IJD
 
 | | The crystal structure of mouse TLR4/MD-2/lipid A complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJC
 
 | | The crystal structure of mouse TLR4/MD-2/neoseptin-3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJB
 
 | | The ligand-free structure of the mouse TLR4/MD-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7C4J
 
 | | Cryo-EM structure of the yeast Swi/Snf complex in a nucleosome free state | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Regulator of Ty1 transposition protein 102, ... | | Authors: | Wang, C.C, Guo, Z.Y, Zhan, X.C, Zhang, X.F. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the yeast Swi/Snf complex in a nucleosome free state
Nat Commun, 2020
|
|
3P2D
 
 | |
3LD2
 
 | |
8H6K
 
 | | Cryo-EM structure of human exon-defined spliceosome in the mature B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6L
 
 | | Cryo-EM structure of human exon-defined spliceosome in the early B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6J
 
 | | Cryo-EM structure of human exon-defined spliceosome in the mature pre-B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6E
 
 | | Cryo-EM structure of human exon-defined spliceosome in the late pre-B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-17 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
