7QHD
 
 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((2-(1H-indol-3-yl)ethyl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | Descriptor: | (3~{S})-1-[[4-[2-(1~{H}-indol-3-yl)ethylcarbamoyl]phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
8I3D
 
 | |
8I39
 
 | | Cryo-EM structure of abscisic acid transporter AtABCG25 with ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABC transporter G family member 25 | | Authors: | Huang, X, Zhang, X, Zhang, P. | | Deposit date: | 2023-01-16 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure and molecular mechanism of abscisic acid transporter ABCG25.
Nat.Plants, 9, 2023
|
|
8I3B
 
 | |
8I3C
 
 | |
8I38
 
 | |
8I3A
 
 | |
3E9G
 
 | | Crystal structure long-form (residue1-124) of Eaf3 chromo domain | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | Deposit date: | 2008-08-22 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
8IAB
 
 | | The Arabidopsis CLCa transporter bound with chloride, ATP and PIP2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Chloride channel protein CLC-a, ... | | Authors: | Yang, Z, Zhang, X, Zhang, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
8IAD
 
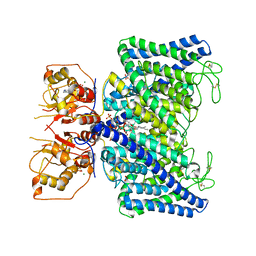 | | The Arabidopsis CLCa transporter bound with nitrate, ATP and PIP2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | Authors: | Yang, Z, Zhang, X, Zhang, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
7ASL
 
 | |
7ASH
 
 | |
8XAT
 
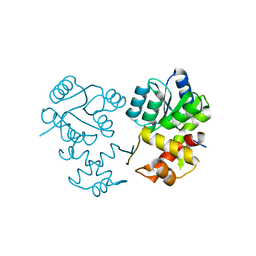 | |
8XAS
 
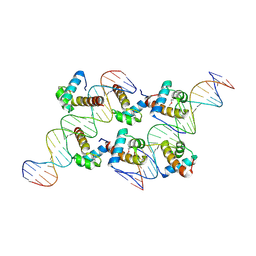 | | Crystal structure of AtARR1-DBD in complex with a DNA fragment | | Descriptor: | DNA (50-MER), Two-component response regulator ARR1 | | Authors: | Li, J.X, Zhou, C.M, zhang, P, Wang, J.W. | | Deposit date: | 2023-12-05 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | The structure of B-ARR reveals the molecular basis of transcriptional activation by cytokinin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4R5J
 
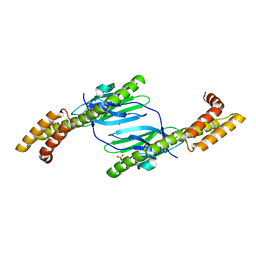 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-A) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, PHOSPHATE ION | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5L
 
 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-C) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, PHOSPHATE ION, ... | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9701 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5G
 
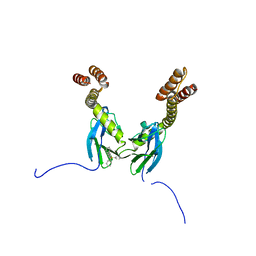 | | Crystal structure of the DnaK C-terminus with the inhibitor PET-16 | | Descriptor: | Chaperone protein DnaK, triphenyl(phenylethynyl)phosphonium | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4501 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5I
 
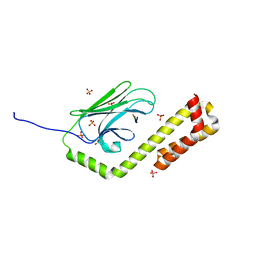 | | Crystal structure of the DnaK C-terminus with the substrate peptide NRLLLTG | | Descriptor: | Chaperone protein DnaK, HSP70/DnaK Substrate Peptide: NRLLLTG, PHOSPHATE ION, ... | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9702 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
8X7T
 
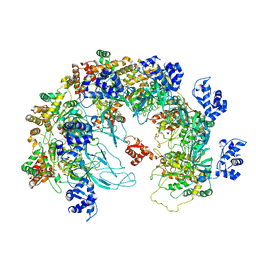 | | MCM in the Apo state. | | Descriptor: | mini-chromosome maintenance complex 3 | | Authors: | Ma, J, Yi, G, Ye, M, MacGregor-Chatwin, C, Sheng, Y, Lu, Y, Li, M, Gilbert, R.J.C, Zhang, P. | | Deposit date: | 2023-11-25 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | MCM in the Apo state
To Be Published
|
|
8X7U
 
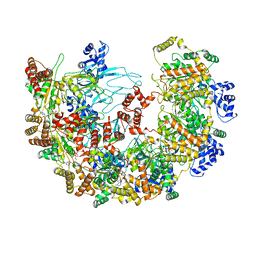 | | MCM in complex with dsDNA in presence of ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, mini-chromosome maintenance complex 3 | | Authors: | Ma, J, Yi, G, Ye, M, MacGregor-Chatwin, C, Sheng, Y, Lu, Y, Li, M, Gilbert, R.J.C, Zhang, P. | | Deposit date: | 2023-11-25 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | MCM in complex with dsDNA in presence of ATP
To Be Published
|
|
8Y7E
 
 | | Cryo-EM Structure of the human minor pre-B complex (pre-precatalytic spliceosome) U12 snRNP part | | Descriptor: | PHD finger-like domain-containing protein 5A, Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, ... | | Authors: | Bai, R, Yuan, M, Zhang, P, Luo, T, Shi, Y, Wan, R. | | Deposit date: | 2024-02-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome.
Science, 383, 2024
|
|
1ZUC
 
 | | Progesterone receptor ligand binding domain in complex with the nonsteroidal agonist tanaproget | | Descriptor: | 5-(4,4-DIMETHYL-2-THIOXO-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-6-YL)-1-METHYL-1H-PYRROLE-2-CARBONITRILE, Progesterone receptor, SULFATE ION | | Authors: | Zhang, Z, Olland, A.M, Zhu, Y, Cohen, J, Berrodin, T, Chippari, S, Appavu, C, Li, S, Wilhem, J, Chopra, R, Fensome, A, Zhang, P, Wrobel, J, Unwalla, R.J, Lyttle, C.R, Winneker, R.C. | | Deposit date: | 2005-05-30 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular and pharmacological properties of a potent and selective novel nonsteroidal progesterone receptor agonist tanaproget
J.Biol.Chem., 280, 2005
|
|
6Q0X
 
 | |
6SKM
 
 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-13,12) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-26 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SLQ
 
 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-12,11) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
