5DP7
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 5 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-methylbutanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
4KYK
 
 | | Crystal structure of mouse glyoxalase I complexed with indomethacin | | Descriptor: | INDOMETHACIN, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhai, J, Yuan, M, Zhang, L, Chen, Y, Zhang, H, Chen, S, Zhao, Y. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Zopolrestat as a human glyoxalase I inhibitor and its structural basis.
Chemmedchem, 8, 2013
|
|
7FHA
 
 | | Crystal structure of the ATP sulfurylase domain of human PAPSS2 in complex with APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthase 2, POTASSIUM ION, ... | | Authors: | Zhang, P, Zhang, L, Zhang, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the substrate recognition mechanism of ATP-sulfurylase domain of human PAPS synthase 2.
Biochem.Biophys.Res.Commun., 586, 2022
|
|
7FH3
 
 | | Crystal structure of the ATP sulfurylase domain of human PAPSS2 | | Descriptor: | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthase 2, SULFATE ION, beta-D-glucopyranose | | Authors: | Zhang, P, Zhang, L, Zhang, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate recognition mechanism of ATP-sulfurylase domain of human PAPS synthase 2.
Biochem.Biophys.Res.Commun., 586, 2022
|
|
6HYH
 
 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis in complex with Beta-D-Fucofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, ZINC ION, beta-D-fucofuranose | | Authors: | Li, M, Mueller, C, Zhang, L, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-10-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
7CAL
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Inwardly Rectifying Potassium Channel KAT1 from Arabidopsis | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Potassium channel KAT1 | | Authors: | Li, S.Y, Yang, F, Sun, D.M, Zhang, Y, Zhang, M.G, Zhou, P, Liu, S.L, Zhang, Y.N, Zhang, L.H, Tian, C.L. | | Deposit date: | 2020-06-09 | | Release date: | 2020-07-29 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the hyperpolarization-activated inwardly rectifying potassium channel KAT1 from Arabidopsis.
Cell Res., 30, 2020
|
|
7C81
 
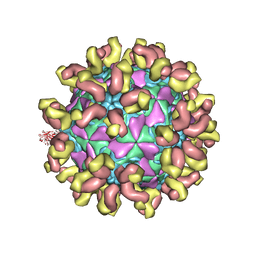 | | E30 F-particle in complex with 6C5 | | Descriptor: | Heavy chain, Light chain, SPHINGOSINE, ... | | Authors: | Wang, K, Zheng, B, Zhang, L, Cui, L, Su, X, Zhang, Q, Guo, Y, Zhu, L, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
8PO8
 
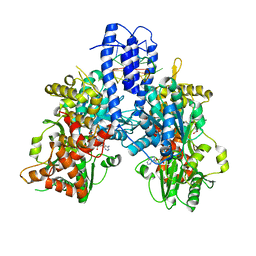 | | Structure of Escherichia coli HrpA in complex with ADP and oligonucleotide poly(dC)11 forming an i-motif | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpA, DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
8PO7
 
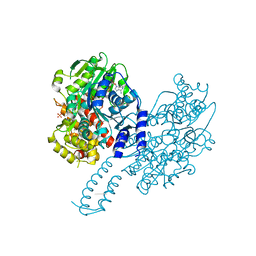 | | Structure of Escherichia coli HrpA in complex with ADP and dinucleotide dCdC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpA, DNA (5'-D(P*CP*C)-3'), ... | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
8PO6
 
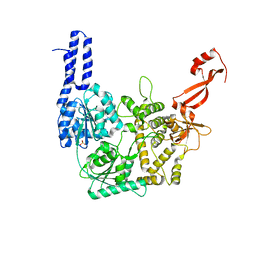 | | Structure of Escherichia coli HrpA apo form | | Descriptor: | ATP-dependent RNA helicase HrpA, PHOSPHATE ION | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
7CDI
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
7CDJ
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1A3 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1A3 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.396 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
7CFM
 
 | | Cryo-EM structure of the P395-bound GPBAR-Gs complex | | Descriptor: | 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5H-pyrido[4,3-d]pyrimidine-4-carboxamide, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
8JFJ
 
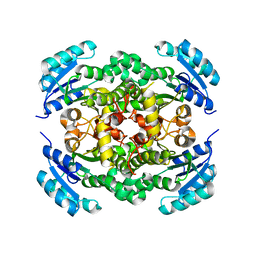 | | Crystal structure of enoyl-ACP reductase FabI from Helicobacter pylori | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Song, W.Y, Zhang, L. | | Deposit date: | 2023-05-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Molecular Basis of Catalysis by SDR Family Members Ketoacyl-ACP Reductase FabG and Enoyl-ACP Reductase FabI in Type-II Fatty Acid Biosynthesis.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
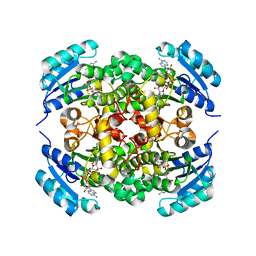
8JFM
 
 | |
8JFG
 
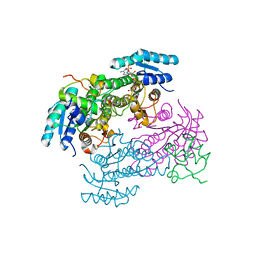 | |
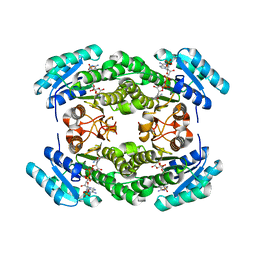
8JFA
 
 | |
8JFH
 
 | |
7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | Descriptor: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
7C80
 
 | | E30 F-particle in complex with 4B10 | | Descriptor: | Heavy chain, Light chain, SPHINGOSINE, ... | | Authors: | Wang, K, Zheng, B, Zhang, L, Cui, L, Su, X, Zhang, Q, Guo, Y, Zhu, L, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
5J19
 
 | | phospho-Pon binding-induced Plk1 dimerization | | Descriptor: | GLYCEROL, Phosphorylated peptide from Partner of Numb, Serine/threonine-protein kinase PLK1 | | Authors: | Zhu, K, Shan, Z, Zhang, L, Wen, W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phospho-Pon Binding-Mediated Fine-Tuning of Plk1 Activity
Structure, 24, 2016
|
|
4KYH
 
 | | Crystal structure of mouse glyoxalase I complexed with zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhai, J, Yuan, M, Zhang, L, Chen, Y, Zhang, H, Chen, S, Zhao, Y. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Zopolrestat as a human glyoxalase I inhibitor and its structural basis.
Chemmedchem, 8, 2013
|
|
3ZWM
 
 | | Crystal structure of ADP ribosyl cyclase complexed with substrate NAD and product cADPR | | Descriptor: | ADP-RIBOSYL CYLCASE, CYCLIC ADENOSINE DIPHOSPHATE-RIBOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZWO
 
 | | Crystal structure of ADP ribosyl cyclase complexed with reaction intermediate | | Descriptor: | 3-(AMINOCARBONYL)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYD ROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2- YL]PYRIDINIUM, ADP-RIBOSYL CYCLASE, GUANOSINE DIPHOSPHATE RIBOSE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZWY
 
 | | Crystal structure of ADP-ribosyl cyclase complexed with 8-bromo-ADP- ribose and cyclic 8-bromo-cyclic-ADP-ribose | | Descriptor: | (2R,3R,4S,5R,13R,14S,15R,16R)-24-amino-18-bromo-3,4,14,15-tetrahydroxy-7,9,11,25,26-pentaoxa-17,19,22-triaza-1-azonia-8 ,10-diphosphapentacyclo[18.3.1.1^2,5^.1^13,16^.0^17,21^]hexacosa-1(24),18,20,22-tetraene-8,10-diolate 8,10-dioxide, ADP-RIBOSYL CYCLASE, [(2R,3S,4R,5R)-5-(6-amino-8-bromo-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4S)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
To be Published
|
|
