3ZDB
 
 | | Structure of E. coli ExoIX in complex with the palindromic 5ov4 DNA oligonucleotide, di-magnesium and potassium | | 分子名称: | 5OV4 DNA, 5'-D(*AP*AP*AP*AP*GP*CP*GP*TP*AP*CP*GP*CP)-3', ACETATE ION, ... | | 著者 | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | 登録日 | 2012-11-26 | | 公開日 | 2013-07-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3ZDE
 
 | | Potassium free structure of E. coli ExoIX | | 分子名称: | PROTEIN XNI | | 著者 | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | 登録日 | 2012-11-26 | | 公開日 | 2013-07-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3ZDC
 
 | | Structure of E. coli ExoIX in complex with the palindromic 5ov4 DNA oligonucleotide, potassium and calcium | | 分子名称: | 5OV4 DNA, 5'-D(*AP*AP*AP*AP*GP*CP*GP*TP*AP*CP*GP*CP)-3', ACETATE ION, ... | | 著者 | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | 登録日 | 2012-11-26 | | 公開日 | 2013-07-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3ZDD
 
 | | Structure of E. coli ExoIX in complex with the palindromic 5ov6 oligonucleotide and potassium | | 分子名称: | 1,2-ETHANEDIOL, 5OV6 DNA, ISOPROPYL ALCOHOL, ... | | 著者 | Flemming, C.S, Hemsworth, G.R, Anstey-Gilbert, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | 登録日 | 2012-11-26 | | 公開日 | 2013-07-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The structure of Escherichia coli ExoIX--implications for DNA binding and catalysis in flap endonucleases.
Nucleic Acids Res., 41, 2013
|
|
3ZD8
 
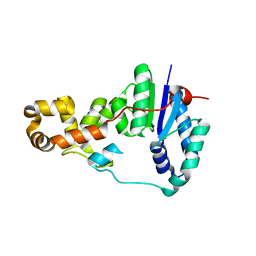 | | Potassium bound structure of E. coli ExoIX in P1 | | 分子名称: | POTASSIUM ION, PROTEIN XNI | | 著者 | Anstey-Gilbert, C.S, Hemsworth, G.R, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | 登録日 | 2012-11-26 | | 公開日 | 2013-07-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
2MOX
 
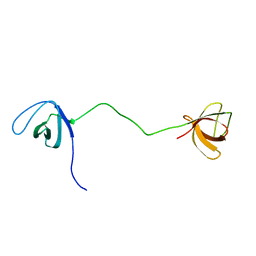 | | solution structure of tandem SH3 domain of Sorbin and SH3 domain-containing protein 1 | | 分子名称: | Sorbin and SH3 domain-containing protein 1 | | 著者 | Zhao, D, Wang, C, Zhang, J, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | 登録日 | 2014-05-07 | | 公開日 | 2014-05-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
8WZQ
 
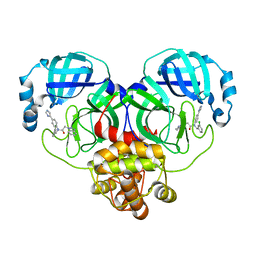 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with CCF0058981 | | 分子名称: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | 著者 | Zou, X.F, Jiang, H.H, Zhou, X.L, Zhang, J, Li, J. | | 登録日 | 2023-11-02 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
8WZP
 
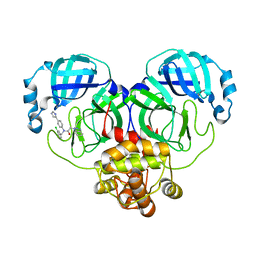 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with CCF0058981 | | 分子名称: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | 著者 | Jiang, H.H, Zou, X.F, Zhou, X.L, Zhang, J, Li, J. | | 登録日 | 2023-11-02 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
5HP4
 
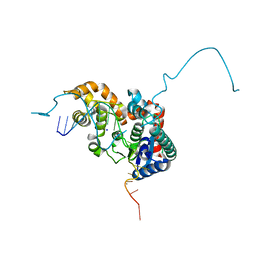 | | Crystal structure bacteriohage T5 D15 flap endonuclease (D155K) pseudo-enzyme-product complex with DNA and metal ions | | 分子名称: | CALCIUM ION, DNA (5'-D(*GP*AP*TP*CP*TP*AP*TP*AP*TP*GP*CP*CP*AP*TP*CP*GP*G)-3'), Exodeoxyribonuclease, ... | | 著者 | Almalki, F.A, Zhang, J, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.A. | | 登録日 | 2016-01-20 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3L5I
 
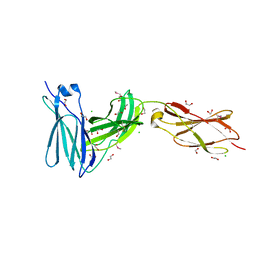 | | Crystal structure of FnIII domains of human GP130 (Domains 4-6) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Interleukin-6 receptor subunit beta | | 著者 | Kershaw, N.J, Zhang, J.-G, Garrett, T.P.J, Czabotar, P.E. | | 登録日 | 2009-12-22 | | 公開日 | 2010-05-12 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes.
J.Biol.Chem., 285, 2010
|
|
8HUW
 
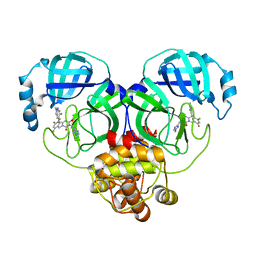 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with S217622 | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Wang, J, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HQG
 
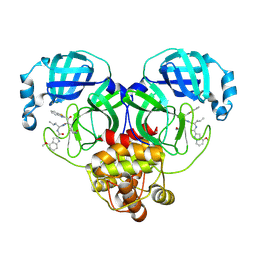 | |
8HVM
 
 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Wang, J, Zhang, J, Li, J. | | 登録日 | 2022-12-27 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
K90R mutant in complex with PF07321332
To Be Published
|
|
6VXN
 
 | |
3MGM
 
 | | Crystal structure of human NUDT16 | | 分子名称: | U8 snoRNA-decapping enzyme | | 著者 | Yan, J, Lu, G, Zhang, J, Qi, J, Li, Z, Gao, F. | | 登録日 | 2010-04-07 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Crystal structure and the template mRNA decapping activity of human NUDT16
To be Published
|
|
6VXM
 
 | |
3L5J
 
 | | Crystal structure of FnIII domains of human GP130 (Domains 4-6) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Interleukin-6 receptor subunit beta | | 著者 | Kershaw, N.J, Zhang, J.-G, Garrett, T.P.J, Czabotar, P.E. | | 登録日 | 2009-12-22 | | 公開日 | 2010-05-12 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.042 Å) | | 主引用文献 | Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes.
J.Biol.Chem., 285, 2010
|
|
8U2B
 
 | |
1XK9
 
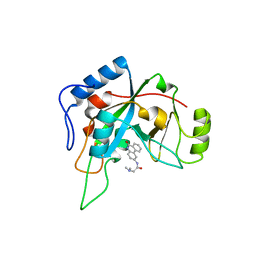 | | Pseudomanas exotoxin A in complex with the PJ34 inhibitor | | 分子名称: | Exotoxin A, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE | | 著者 | Yates, S.P, Taylor, P.J, Joergensen, R, Ferrraris, D, Zhang, J, Andersen, G.R, Merrill, A.R. | | 登録日 | 2004-09-28 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-function analysis of water-soluble inhibitors of the catalytic domain of exotoxin A from Pseudomonas aeruginosa
BIOCHEM.J., 385, 2005
|
|
3J32
 
 | | An asymmetric unit map from electron cryo-microscopy of Haliotis diversicolor molluscan hemocyanin isoform 1 (HdH1) | | 分子名称: | Hemocyanin isoform 1 | | 著者 | Zhang, Q, Dai, X, Cong, Y, Zhang, J, Chen, D.-H, Dougherty, M, Wang, J, Ludtke, S, Schmid, M.F, Chiu, W. | | 登録日 | 2013-02-20 | | 公開日 | 2013-04-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Cryo-EM structure of a molluscan hemocyanin suggests its allosteric mechanism.
Structure, 21, 2013
|
|
2L89
 
 | |
2LNW
 
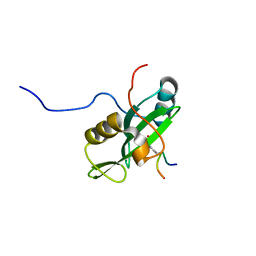 | | Identification and structural basis for a novel interaction between Vav2 and Arap3 | | 分子名称: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3, Guanine nucleotide exchange factor VAV2 | | 著者 | Wu, B, Zhang, J, Wu, J, Shi, Y. | | 登録日 | 2012-01-05 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
5Z6P
 
 | | The crystal structure of an agarase, AgWH50C | | 分子名称: | B-agarase | | 著者 | Mao, X, Zhou, J, Zhang, P, Zhang, L, Zhang, J, Li, Y. | | 登録日 | 2018-01-24 | | 公開日 | 2019-01-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.061 Å) | | 主引用文献 | Structure-based design of agarase AgWH50C from Agarivorans gilvus WH0801 to enhance thermostability.
Appl. Microbiol. Biotechnol., 103, 2019
|
|
7XF3
 
 | | The structure of HLA-B*1501/BM58-66AF9 | | 分子名称: | 9-mer peptide from Matrix protein 1, Beta-2-microglobulin, MHC class I antigen | | 著者 | Zhao, Y.Z, Xiao, W.L, Wu, Y.N, Fan, W.F, Yue, C, Zhang, Q.X, Zhang, D.N, Yuan, X.J, Yao, S.J, Liu, S, Li, M, Wang, P.Y, Zhang, H.J, Zhang, J, Zhao, M, Zheng, X.Q, Liu, W.J, Gao, G.F, Liu, W.L. | | 登録日 | 2022-03-31 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Parallel T Cell Immunogenic Regions in Influenza B and A Viruses with Distinct Nuclear Export Signal Functions: The Balance between Viral Life Cycle and Immune Escape.
J Immunol., 210, 2023
|
|
2LNX
 
 | |
