6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6M0K
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11b | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
8I16
 
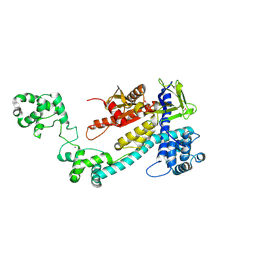 | | Crystal structure of the selenomethionine (SeMet)-derived Cas12g (D513A) mutant | | Descriptor: | Cas12g, ZINC ION | | Authors: | Zhang, B, Chen, J, Ye, Y.M, OuYang, S.Y. | | Deposit date: | 2023-01-12 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural transitions upon guide RNA binding and their importance in Cas12g-mediated RNA cleavage.
Plos Genet., 19, 2023
|
|
5DE9
 
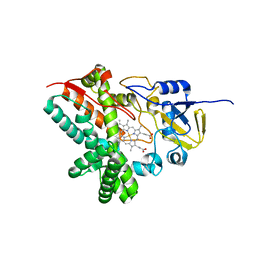 | | The role of Ile87 of CYP158A2 in oxidative coupling reaction | | Descriptor: | Biflaviolin synthase CYP158A2, PROTOPORPHYRIN IX CONTAINING FE, SPERMINE | | Authors: | Zhao, B. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The role of Ile87 of CYP158A2 in oxidative coupling reaction.
Arch. Biochem. Biophys., 518, 2012
|
|
4IMA
 
 | | The structure of C436M-hLPYK in complex with Citrate/Mn/ATP/Fru-1,6-BP | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE, ... | | Authors: | Zhang, B, Holyoak, T, Fenton, A.W, Tang, Q.L, Prasannan, C.B, Deng, J.P. | | Deposit date: | 2013-01-02 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Energetic Coupling between an Oxidizable Cysteine and the Phosphorylatable N-Terminus of Human Liver Pyruvate Kinase.
Biochemistry, 52, 2013
|
|
7SUX
 
 | |
7SV0
 
 | |
7SUZ
 
 | |
3E5A
 
 | | Crystal structure of Aurora A in complex with VX-680 and TPX2 | | Descriptor: | CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, SULFATE ION, Serine/threonine-protein kinase 6, ... | | Authors: | Zhao, B, Smallwood, A, Lai, Z. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modulation of kinase-inhibitor interactions by auxiliary protein binding: crystallography studies on Aurora A interactions with VX-680 and with TPX2.
Protein Sci., 17, 2008
|
|
4XO7
 
 | | Crystal structure of human 3-alpha hydroxysteroid dehydrogenase type 3 in complex with NADP+, 5alpha-androstan-3,17-dione and (3beta, 5alpha)-3-hydroxyandrostan-17-one | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, Aldo-keto reductase family 1 member C2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Human 3 alpha-hydroxysteroid dehydrogenase type 3: structural clues of 5 alpha-DHT reverse binding and enzyme down-regulation decreasing MCF7 cell growth.
Biochem.J., 473, 2016
|
|
6S7V
 
 | | Lipoteichoic acids flippase LtaA | | Descriptor: | MFS transporter | | Authors: | Zhang, B, Perez, C. | | Deposit date: | 2019-07-06 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Structure of a proton-dependent lipid transporter involved in lipoteichoic acids biosynthesis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4XO6
 
 | | Crystal structure of human 3-alpha hydroxysteroid dehydrogenase type 3 in complex with NADP+, 5alpha-androstan-3,17-dione and (3beta, 5alpha)-3-hydroxyandrostan-17-one | | Descriptor: | (3Beta,5alpha)-3-Hydroxyandrostan-17-one, 1,2-ETHANEDIOL, 5ALPHA-ANDROSTAN-3,17-DIONE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Human 3 alpha-hydroxysteroid dehydrogenase type 3: structural clues of 5 alpha-DHT reverse binding and enzyme down-regulation decreasing MCF7 cell growth.
Biochem.J., 473, 2016
|
|
6IV8
 
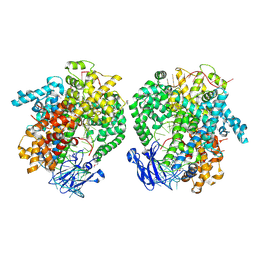 | | the selenomethionine(SeMet)-derived Cas13d binary complex | | Descriptor: | MAGNESIUM ION, RNA (51-MER), RNA (53-MER), ... | | Authors: | Zhang, B, Ye, Y.M, Ye, W.W, OuYang, S.Y. | | Deposit date: | 2018-12-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d.
Nat Commun, 10, 2019
|
|
6IV9
 
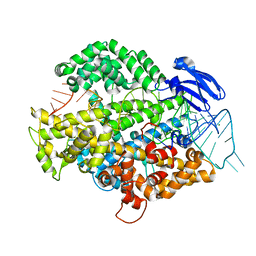 | | the Cas13d binary complex | | Descriptor: | Cas13d, MAGNESIUM ION, crRNA (50-MER) | | Authors: | Zhang, B, Ye, Y.M, Ye, W.W, OuYang, S.Y. | | Deposit date: | 2018-12-02 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d.
Nat Commun, 10, 2019
|
|
2D09
 
 | |
2D0E
 
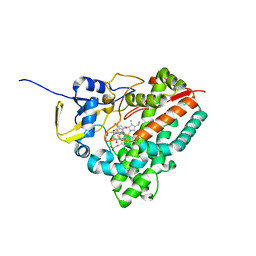 | | Substrate assited in Oxygen Activation in Cytochrome P450 158A2 | | Descriptor: | 2-HYDROXYNAPHTHOQUINONE, PROTOPORPHYRIN IX CONTAINING FE, putative cytochrome P450 | | Authors: | Zhao, B, Waterman, M.R. | | Deposit date: | 2005-08-02 | | Release date: | 2005-10-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Role of active site water molecules and substrate hydroxyl groups in oxygen activation by cytochrome P450 158A2: a new mechanism of proton transfer
J.Biol.Chem., 280, 2005
|
|
1NVS
 
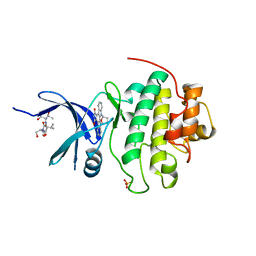 | | The Complex Structure Of Checkpoint Kinase Chk1/SB218078 | | Descriptor: | Peptide ASVSA, REL-(9R,12S)-9,10,11,12-TETRAHYDRO-9,12-EPOXY-1H-DIINDOLO[1,2,3-FG:3',2',1'-KL]PYRROLO[3,4-I][1,6]BENZODIAZOCINE-1,3(2H)-DIONE, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
7KPT
 
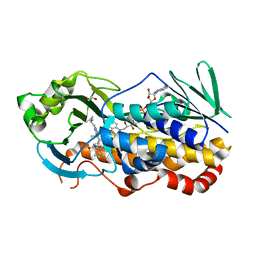 | | Crystal structure of CtdE in complex with FAD and substrate 4 | | Descriptor: | (6aR,7aS,11S,13aS)-6,6,11-trimethyl-4-(3-methylbut-2-en-1-yl)-6,6a,7,8,9,10,11,14-octahydro-5H,13H-13a,7a-(epiminomethano)quinolizino[2,3-b]carbazol-16-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Zhao, B, Hu, L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis of the stereoselective formation of the spirooxindole ring in the biosynthesis of citrinadins.
Nat Commun, 12, 2021
|
|
7KPQ
 
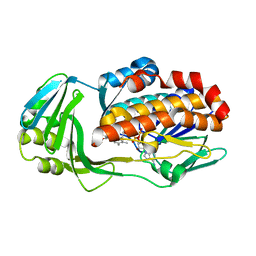 | | Crystal structure of CtdE in complex with FAD | | Descriptor: | FAD-dependent monooxygenase CtdE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhao, B, Hu, L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the stereoselective formation of the spirooxindole ring in the biosynthesis of citrinadins.
Nat Commun, 12, 2021
|
|
1NVQ
 
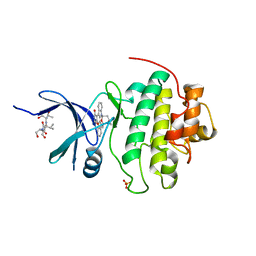 | | The Complex Structure Of Checkpoint Kinase Chk1/UCN-01 | | Descriptor: | 7-HYDROXYSTAUROSPORINE, Peptide ASVSA, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
1NVR
 
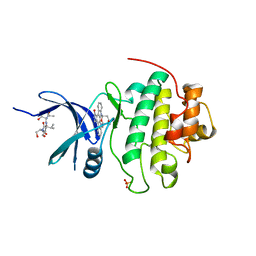 | | The Complex Structure Of Checkpoint Kinase Chk1/Staurosporine | | Descriptor: | Peptide ASVSA, STAUROSPORINE, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
5IF4
 
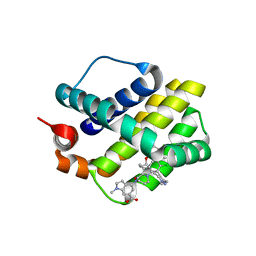 | | Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) inhibitors using Structure-Based Design | | Descriptor: | 4-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-6-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Discovery and biological characterization of potent myeloid cell leukemia-1 inhibitors.
FEBS Lett., 591, 2017
|
|
5IEZ
 
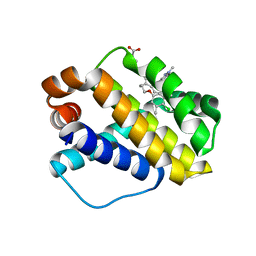 | | Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) inhibitors using Structure-Based Design | | Descriptor: | 3-({6-chloro-3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-1H-indole-2-carbonyl}amino)benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and biological characterization of potent myeloid cell leukemia-1 inhibitors.
FEBS Lett., 591, 2017
|
|
5JEK
 
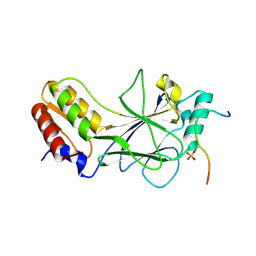 | | Phosphorylated MAVS in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, MAVS peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEM
 
 | | Complex of IRF-3 with CBP | | Descriptor: | CREB-binding protein, Interferon regulatory factor 3 | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
