3NGY
 
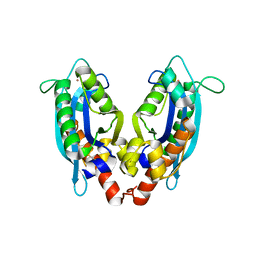 | | Crystal structure of RNase T (E92G mutant) | | Descriptor: | COBALT (II) ION, Ribonuclease T, his tag sequence | | Authors: | Hsiao, Y.-Y, Yuan, H.S. | | Deposit date: | 2010-06-14 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis for RNA trimming by RNase T in stable RNA 3'-end maturation
Nat.Chem.Biol., 7, 2011
|
|
3NGZ
 
 | |
3NH1
 
 | |
3NH2
 
 | |
3NH0
 
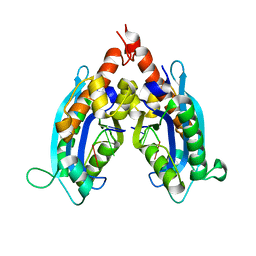 | |
3KRN
 
 | | Crystal Structure of C. elegans cell-death-related nuclease 5(CRN-5) | | Descriptor: | Protein C14A4.5, confirmed by transcript evidence | | Authors: | Yang, C.-C, Wang, Y.-T, Hsiao, Y.-Y, Doudeva, L.G, Chow, S.Y, Yuan, H.S. | | Deposit date: | 2009-11-19 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.918 Å) | | Cite: | Structural and biochemical characterization of CRN-5 and Rrp46: an exosome component participating in apoptotic DNA degradation
Rna, 16, 2010
|
|
1ETQ
 
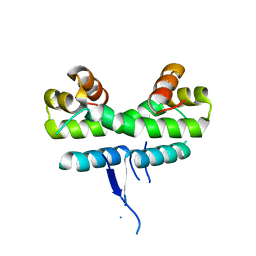 | |
1ETV
 
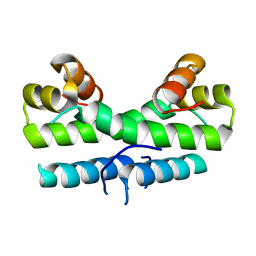 | |
1ETW
 
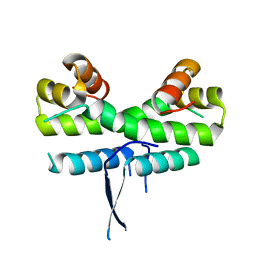 | |
5GKC
 
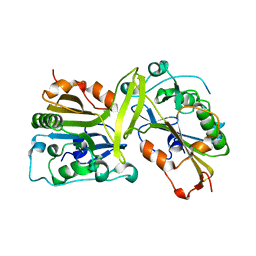 | | The crystal structure of the CPS-6 H148A/F122A | | Descriptor: | Endonuclease G, mitochondrial | | Authors: | Lin, J.L, Yuan, H.S. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Crystal structure of endonuclease G in complex with DNA reveals how it nonspecifically degrades DNA as a homodimer.
Nucleic Acids Res., 44, 2016
|
|
1ETX
 
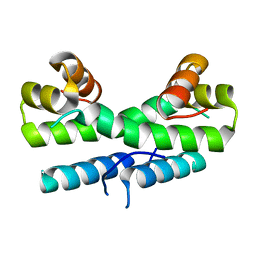 | |
1ETK
 
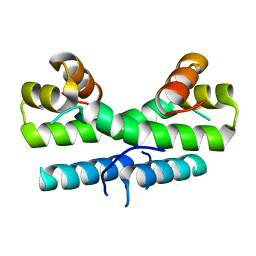 | |
1ETY
 
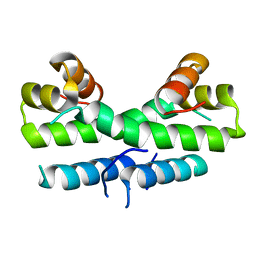 | |
1CEI
 
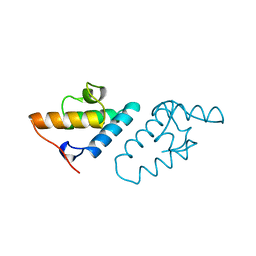 | | STRUCTURE DETERMINATION OF THE COLICIN E7 IMMUNITY PROTEIN (IMME7) THAT BINDS SPECIFICALLY TO THE DNASE-TYPE COLICIN E7 AND INHIBITS ITS BACTERIOCIDAL ACTIVITY | | Descriptor: | COLICIN E7 IMMUNITY PROTEIN | | Authors: | Chak, K.-F, Safo, M.K, Ku, W.-Y, Hsieh, S.-Y, Yuan, H.S. | | Deposit date: | 1996-03-19 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the immunity protein of colicin E7 suggests a possible colicin-interacting surface.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1ETO
 
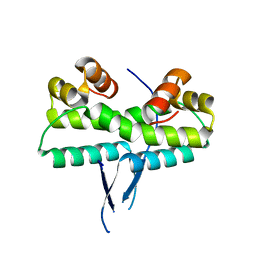 | |
1F36
 
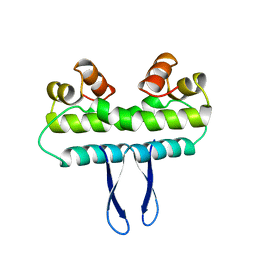 | |
5ZKI
 
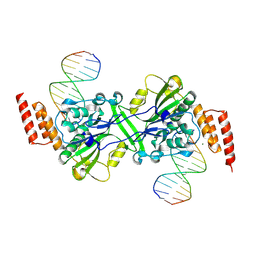 | | Human EXOG-H140A in complex with duplex DNA | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*GP*GP*AP*TP*AP*TP*CP*CP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Wu, C.C, Lin, J.L.J, Yuan, H.S. | | Deposit date: | 2018-03-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | A unique exonuclease ExoG cleaves between RNA and DNA in mitochondrial DNA replication.
Nucleic Acids Res., 47, 2019
|
|
5ZKJ
 
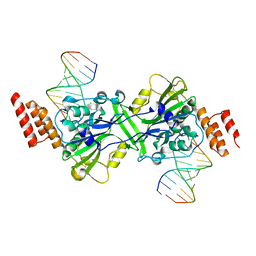 | | Human EXOG-H140A in complex with RNA/DNA hybrid duplex | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*GP*AP*CP*AP*TP*CP*CP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Wu, C.C, Lin, J.L.J, Yuan, H.S. | | Deposit date: | 2018-03-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | A unique exonuclease ExoG cleaves between RNA and DNA in mitochondrial DNA replication.
Nucleic Acids Res., 47, 2019
|
|
7CEI
 
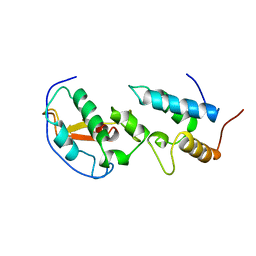 | | THE ENDONUCLEASE DOMAIN OF COLICIN E7 IN COMPLEX WITH ITS INHIBITOR IM7 PROTEIN | | Descriptor: | PROTEIN (COLICIN E7 IMMUNITY PROTEIN), ZINC ION | | Authors: | Ko, T.P, Liao, C.C, Ku, W.Y, Chak, K.F, Yuan, H.S. | | Deposit date: | 1998-09-17 | | Release date: | 1999-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein.
Structure Fold.Des., 7, 1999
|
|
7DA6
 
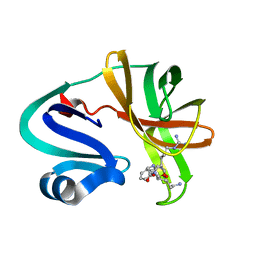 | |
7X9D
 
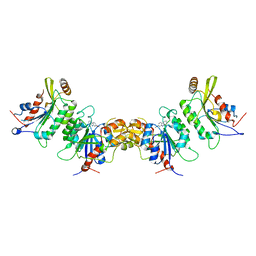 | | DNMT3B in complex with harmine | | Descriptor: | 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B | | Authors: | Cho, C.-C, Yuan, H.S. | | Deposit date: | 2022-03-15 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Mechanistic Insights into Harmine-Mediated Inhibition of Human DNA Methyltransferases and Prostate Cancer Cell Growth.
Acs Chem.Biol., 18, 2023
|
|
7W1R
 
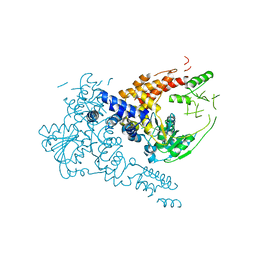 | | Crystal structure of human Suv3 monomer | | Descriptor: | ATP-dependent RNA helicase SUPV3L1, mitochondrial | | Authors: | Jain, M, Yuan, H.S. | | Deposit date: | 2021-11-20 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dimeric assembly of human Suv3 helicase promotes its RNA unwinding function in mitochondrial RNA degradosome for RNA decay.
Protein Sci., 31, 2022
|
|
6J80
 
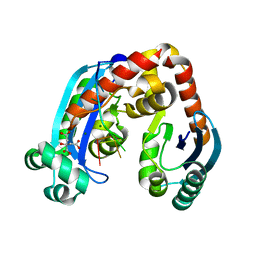 | | Human mitochondrial Oligoribonuclease in complex with poly-dT DNA | | Descriptor: | CITRIC ACID, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Chu, L.Y, Agrawal, S, Yuan, H.S. | | Deposit date: | 2019-01-18 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | Structural insights into nanoRNA degradation by human Rexo2.
Rna, 25, 2019
|
|
6J7Z
 
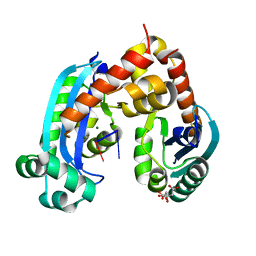 | | Human mitochondrial Oligoribonuclease in complex with RNA | | Descriptor: | CITRIC ACID, MAGNESIUM ION, Oligoribonuclease, ... | | Authors: | Chu, L.Y, Agrawal, S, Yuan, H.S. | | Deposit date: | 2019-01-18 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural insights into nanoRNA degradation by human Rexo2.
Rna, 25, 2019
|
|
6KDB
 
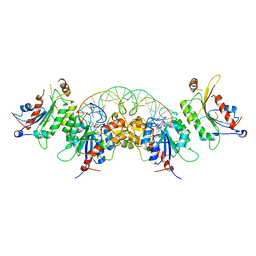 | | Crystal structure of human DNMT3B-DNMT3L in complex with DNA containing CpGpT site | | Descriptor: | DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Lin, C.-C, Chen, Y.-P, Yang, W.-Z, Shen, C.-K, Yuan, H.S. | | Deposit date: | 2019-07-01 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.862 Å) | | Cite: | Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B.
Nucleic Acids Res., 48, 2020
|
|
