4XB7
 
 | | Crystal structure of Dscam1 isoform 4.4, N-terminal four Ig domains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.4, ... | | 著者 | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | 登録日 | 2014-12-16 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (4.004 Å) | | 主引用文献 | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9G
 
 | | Crystal structure of Dscam1 isoform 6.44, N-terminal four Ig domains | | 分子名称: | Down Syndrome Cell Adhesion Molecule isoform 6.44, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | 登録日 | 2014-12-11 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.403 Å) | | 主引用文献 | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9I
 
 | | Crystal structure of Dscam1 isoform 9.44, N-terminal four Ig domains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule, isoform 9.44, ... | | 著者 | Chen, Q, Yu, Y, Li, S.A, cheng, L. | | 登録日 | 2014-12-11 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.904 Å) | | 主引用文献 | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4XB8
 
 | | Crystal structure of Dscam1 isoform 9.44, N-terminal four Ig domains (with zinc) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule, ... | | 著者 | Chen, Q, Yu, Y, Li, S.A, cheng, L. | | 登録日 | 2014-12-16 | | 公開日 | 2015-12-16 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.202 Å) | | 主引用文献 | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
3N6N
 
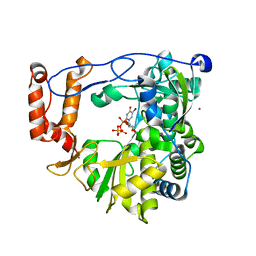 | | crystal structure of EV71 RdRp in complex with Br-UTP | | 分子名称: | 5-bromouridine 5'-(tetrahydrogen triphosphate), NICKEL (II) ION, RNA-dependent RNA polymerase | | 著者 | Wu, Y, Lou, Z.Y, Miao, Y, Yu, Y, Rao, Z.H. | | 登録日 | 2010-05-26 | | 公開日 | 2011-06-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China.
Protein Cell, 1, 2010
|
|
3N6M
 
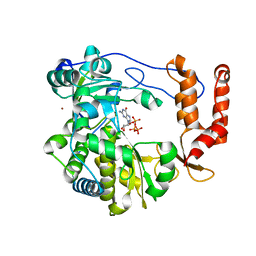 | | Crystal structure of EV71 RdRp in complex with GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, NICKEL (II) ION, RNA-dependent RNA polymerase | | 著者 | Wu, Y, Lou, Z.Y, Miao, Y, Yu, Y, Rao, Z.H. | | 登録日 | 2010-05-26 | | 公開日 | 2011-06-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China.
Protein Cell, 1, 2010
|
|
3C91
 
 | | Thermoplasma acidophilum 20S proteasome with an open gate | | 分子名称: | Proteasome subunit alpha, Proteasome subunit beta | | 著者 | Rabl, J, Smith, D.M, Yu, Y, Chang, S.C, Goldberg, A.L, Cheng, Y. | | 登録日 | 2008-02-14 | | 公開日 | 2008-08-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases.
Mol.Cell, 30, 2008
|
|
3C92
 
 | | Thermoplasma acidophilum 20S proteasome with a closed gate | | 分子名称: | Proteasome subunit alpha, Proteasome subunit beta | | 著者 | Rabl, J, Smith, D.M, Yu, Y, Chang, S.C, Goldberg, A.L, Cheng, Y. | | 登録日 | 2008-02-14 | | 公開日 | 2008-08-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases.
Mol.Cell, 30, 2008
|
|
1AYG
 
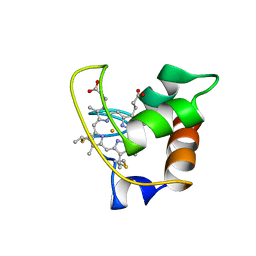 | | SOLUTION STRUCTURE OF CYTOCHROME C-552, NMR, 20 STRUCTURES | | 分子名称: | CYTOCHROME C-552, HEME C | | 著者 | Hasegawa, J, Yoshida, T, Yamazaki, T, Sambongi, Y, Yu, Y, Igarashi, Y, Kodama, T, Yamazaki, K, Hakusui, H, Kyogoku, Y, Kobayashi, Y. | | 登録日 | 1997-11-04 | | 公開日 | 1998-11-25 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of thermostable cytochrome c-552 from Hydrogenobacter thermophilus determined by 1H-NMR spectroscopy.
Biochemistry, 37, 1998
|
|
5BOA
 
 | | Crystal Structure of the Meningitis Pathogen Streptococcus suis adhesion Fhb bound to the disaccharide receptor Gb2 | | 分子名称: | Translation initiation factor 2 (IF-2 GTPase), alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose | | 著者 | Zhang, C, Yu, Y, Yang, M, Jiang, Y. | | 登録日 | 2015-05-27 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.708 Å) | | 主引用文献 | Structural basis of the interaction between the meningitis pathogen Streptococcus suis adhesin Fhb and its human receptor.
Febs Lett., 590, 2016
|
|
7Y4X
 
 | |
5C8Y
 
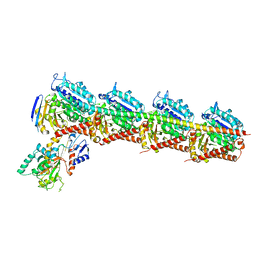 | | Crystal structure of T2R-TTL-Plinabulin complex | | 分子名称: | (3Z,6Z)-3-benzylidene-6-[(5-tert-butyl-1H-imidazol-4-yl)methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | 登録日 | 2015-06-26 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.594 Å) | | 主引用文献 | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
5CA1
 
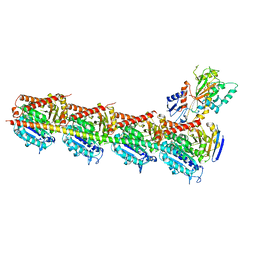 | | Crystal structure of T2R-TTL-Nocodazole complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | 著者 | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | 登録日 | 2015-06-29 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
5CA0
 
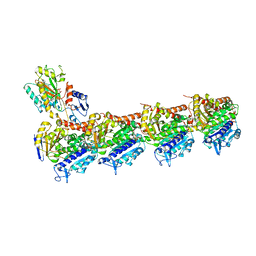 | | Crystal structure of T2R-TTL-Lexibulin complex | | 分子名称: | 1-ethyl-3-[2-methoxy-4-(5-methyl-4-{[(1S)-1-(pyridin-3-yl)butyl]amino}pyrimidin-2-yl)phenyl]urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | 登録日 | 2015-06-29 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
5CB4
 
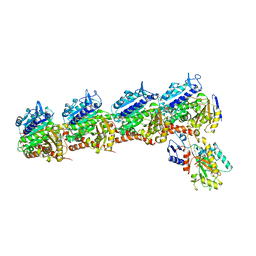 | | Crystal structure of T2R-TTL-Tivantinib complex | | 分子名称: | (3R,4R)-3-(5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1-yl)-4-(1H-indol-3-yl)pyrrolidine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | 登録日 | 2015-06-30 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.193 Å) | | 主引用文献 | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | 著者 | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | 登録日 | 2015-04-13 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
5EZY
 
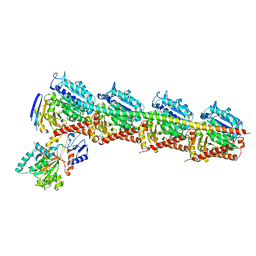 | | Crystal structure of T2R-TTL-taccalonolide AJ complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | 登録日 | 2015-11-27 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Mechanism of microtubule stabilization by taccalonolide AJ
Nat Commun, 8, 2017
|
|
5BOB
 
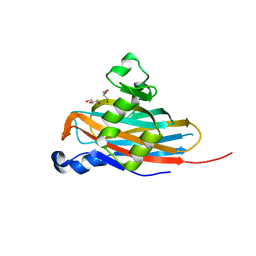 | | Crystal Structure of the Meningitis Pathogen Streptococcus suis adhesion Fhb | | 分子名称: | GLYCEROL, Translation initiation factor 2 (IF-2 GTPase) | | 著者 | Jiang, Y, Zhang, C, Yu, Y. | | 登録日 | 2015-05-27 | | 公開日 | 2015-11-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Expression, purification, crystallization and structure determination of the N terminal domain of Fhb, a factor H binding protein from Streptococcus suis.
Biochem.Biophys.Res.Commun., 466, 2015
|
|
7XRJ
 
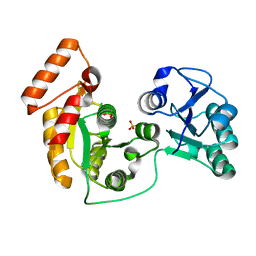 | | crystal structure of N-acetyltransferase DgcN-25328 | | 分子名称: | Putative NAD-dependent epimerase/dehydratase family protein, SULFATE ION | | 著者 | Zhang, Y.Z, Yu, Y, Cao, H.Y, Chen, X.L, Wang, P. | | 登録日 | 2022-05-10 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Novel D-glutamate catabolic pathway in marine Proteobacteria and halophilic archaea.
Isme J, 17, 2023
|
|
7Y54
 
 | |
7Y95
 
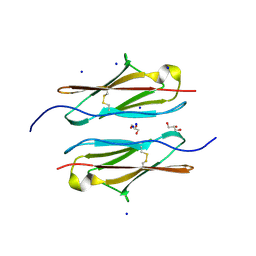 | | Crystal structure of sDscam Ig1 domain, isoform beta6v2 | | 分子名称: | Dscam, GLYCEROL, SODIUM ION | | 著者 | Chen, Q, Yu, Y, Cheng, J. | | 登録日 | 2022-06-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y6E
 
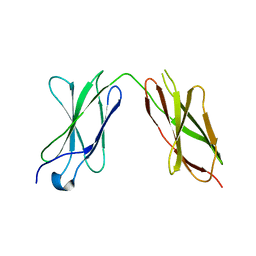 | |
7Y5J
 
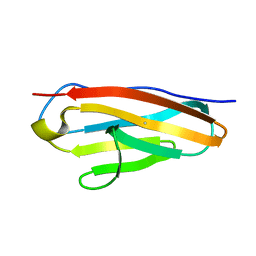 | |
7Y6O
 
 | |
7Y8I
 
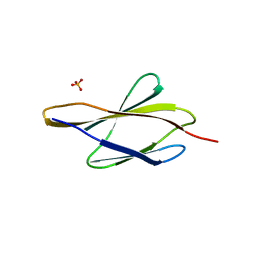 | | Crystal structure of sDscam FNIII3 domain, isoform alpha7 | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dscam, ... | | 著者 | Chen, Q, Yu, Y, Cheng, J. | | 登録日 | 2022-06-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
