6WTP
 
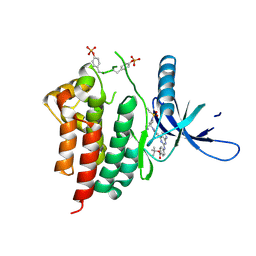 | | Human JAK2 JH1 domain in complex with PROTAC-intermediate linker handle 3 | | Descriptor: | GLYCEROL, Tyrosine-protein kinase JAK2, tert-butyl 4-[(4-{1-[3-(cyanomethyl)-1-(ethylsulfonyl)azetidin-3-yl]-1H-pyrazol-4-yl}-7H-pyrrolo[2,3-d]pyrimidin-2-yl)amino]benzoate | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTO
 
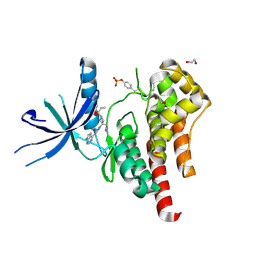 | | Human JAK2 JH1 domain in complex with Baricitinib | | Descriptor: | 1,2-ETHANEDIOL, Baricitinib, Tyrosine-protein kinase JAK2 | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTQ
 
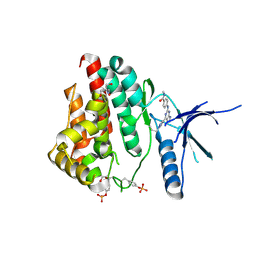 | | Human JAK2 JH1 domain in complex with PROTAC-intermediate linker handle 4 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-4-{[4-(1-propyl-1H-pyrazol-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl]amino}benzamide, ... | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79968476 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTN
 
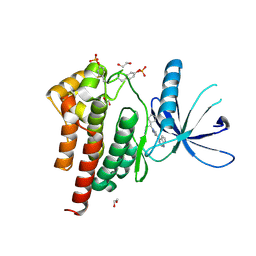 | | Human JAK2 JH1 domain in complex with Ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, 1,2-ETHANEDIOL, Tyrosine-protein kinase JAK2 | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
3DH8
 
 | | Structure of Pseudomonas Quinolone Signal Response Protein PqsE | | Descriptor: | FE (III) ION, Uncharacterized protein PA1000, bis(4-nitrophenyl) hydrogen phosphate | | Authors: | Yu, S, Blankenfeldt, W. | | Deposit date: | 2008-06-17 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein.
Biochemistry, 48, 2009
|
|
6KDD
 
 | | endoglucanase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase, GLYCEROL, ... | | Authors: | Yu, S, Liuqing, C. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | crystal structure of an endoglucanase from Fervidobacterium pennivorans DSM9078
To Be Published
|
|
3P2F
 
 | | Crystal structure of TofI in an apo form | | Descriptor: | AHL synthase | | Authors: | Yu, S, Rhee, S. | | Deposit date: | 2010-10-02 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule inhibitor binding to an N-acyl-homoserine lactone synthase
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3P2H
 
 | |
3K2D
 
 | |
2Q0I
 
 | | Structure of Pseudomonas Quinolone Signal Response Protein PqsE | | Descriptor: | BENZOIC ACID, FE (III) ION, Quinolone signal response protein | | Authors: | Yu, S, Jensen, V, Feldmann, I, Haussler, S, Blankenfeldt, W. | | Deposit date: | 2007-05-22 | | Release date: | 2008-06-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein.
Biochemistry, 48, 2009
|
|
2Q0J
 
 | | Structure of Pseudomonas Quinolone Signal Response Protein PqsE | | Descriptor: | BENZOIC ACID, FE (III) ION, Quinolone signal response protein | | Authors: | Yu, S, Jensen, V, Feldmann, I, Haussler, S, Blankenfeldt, W. | | Deposit date: | 2007-05-22 | | Release date: | 2008-06-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein.
Biochemistry, 48, 2009
|
|
4KL0
 
 | | Crystal structure of the effector protein XOO4466 | | Descriptor: | CALCIUM ION, Putative uncharacterized protein | | Authors: | Yu, S, Rhee, S. | | Deposit date: | 2013-05-07 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure of the effector protein XOO4466 from Xanthomonas oryzae
J.Struct.Biol., 184, 2013
|
|
4P5F
 
 | | The crystal structure of type III effector protein XopQ complexed with adenosine diphosphate ribose | | Descriptor: | CALCIUM ION, Inosine-uridine nucleoside N-ribohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Yu, S, Hwang, I, Rhee, S. | | Deposit date: | 2014-03-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of type III effector protein XopQ from Xanthomonas oryzae complexed with adenosine diphosphate ribose.
Proteins, 82, 2014
|
|
6KDC
 
 | |
2X2I
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
2X2H
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase | | Descriptor: | ACETATE ION, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
2X2J
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha- 1,4-glucan lyase with deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
3DSS
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) | | Descriptor: | CALCIUM ION, Geranylgeranyl transferase type-2 subunit alpha, Geranylgeranyl transferase type-2 subunit beta, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3DSX
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with di-prenylated peptide Ser-Cys(GG)-Ser-Cys(GG) derivated from Rab7 | | Descriptor: | CALCIUM ION, GERAN-8-YL GERAN, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3DSW
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with mono-prenylated peptide Ser-Cys(GG)-Ser-Cys derivated from Rab7 | | Descriptor: | CALCIUM ION, GERAN-8-YL GERAN, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3DSU
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with farnesyl pyrophosphate | | Descriptor: | CALCIUM ION, FARNESYL DIPHOSPHATE, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3DST
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with geranylgeranyl pyrophosphate | | Descriptor: | CALCIUM ION, GERANYLGERANYL DIPHOSPHATE, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3DSV
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with mono-prenylated peptide Ser-Cys-Ser-Cys(GG) derivated from Rab7 | | Descriptor: | CALCIUM ION, GERAN-8-YL GERAN, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3SK2
 
 | |
3SK1
 
 | |
