1IQU
 
 | | Crystal structure of photolyase-thymine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, THYMINE, ... | | Authors: | Komori, H, Masui, R, Kuramitsu, S, Yokoyama, S, Shibata, T, Inoue, Y, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-08-03 | | Release date: | 2002-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermostable DNA photolyase: pyrimidine-dimer recognition mechanism.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IV1
 
 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6KS0
 
 | | Crystal structure of the human adiponectin receptor 1 | | Descriptor: | Adiponectin receptor protein 1, PHOSPHATE ION, The heavy chain variable domain (Antibody), ... | | Authors: | Tanabe, H, Fujii, Y, Nakamura, Y, Hosaka, T, Okada-Iwabu, M, Iwabu, M, Kimura-Someya, T, Shirouzu, M, Yamauchi, T, Kadowaki, T, Yokoyama, S. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Human adiponectin receptor AdipoR1 assumes closed and open structures.
Commun Biol, 3, 2020
|
|
6KS1
 
 | | Crystal structure of the human adiponectin receptor 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Tanabe, H, Fujii, Y, Nakamura, Y, Hosaka, T, Okada-Iwabu, M, Iwabu, M, Kimura-Someya, T, Shirouzu, M, Yamauchi, T, Kadowaki, T, Yokoyama, S. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human adiponectin receptor AdipoR1 assumes closed and open structures.
Commun Biol, 3, 2020
|
|
6KRZ
 
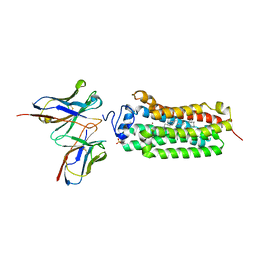 | | Crystal structure of the human adiponectin receptor 1 D208A mutant | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 1, OLEIC ACID, ... | | Authors: | Tanabe, H, Fujii, Y, Okada-Iwabu, M, Iwabu, M, Yamauchi, T, Kadowaki, T, Yokoyama, S. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Human adiponectin receptor AdipoR1 assumes closed and open structures.
Commun Biol, 3, 2020
|
|
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | Descriptor: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | Authors: | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | Deposit date: | 2001-09-27 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
5XAX
 
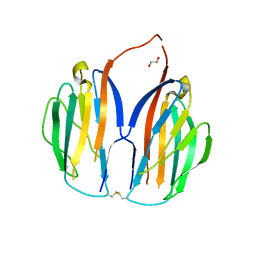 | |
5XAW
 
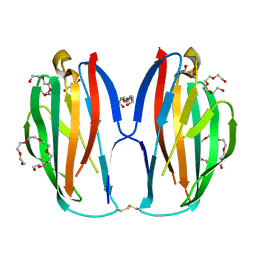 | | Parallel homodimer structures of voltage-gated sodium channel beta4 for cell-cell adhesion | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GLYCEROL, Sodium channel subunit beta-4, ... | | Authors: | Shimizu, H, Yokoyama, S. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Parallel homodimer structures of the extracellular domains of the voltage-gated sodium channel beta 4 subunit explain its role in cell-cell adhesion
J. Biol. Chem., 292, 2017
|
|
1IV0
 
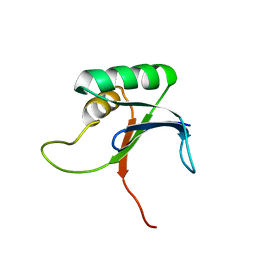 | |
1IYF
 
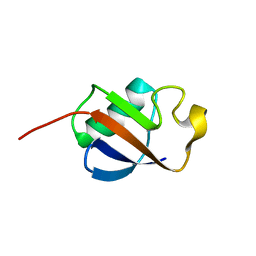 | | Solution structure of ubiquitin-like domain of human parkin | | Descriptor: | parkin | | Authors: | Sakata, E, Yamaguchi, Y, Kurimoto, E, Kikuchi, J, Yokoyama, S, Kawahara, H, Yokosawa, H, Hattori, N, Mizuno, Y, Tanaka, K, Kato, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-13 | | Release date: | 2003-03-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain
EMBO REP., 4, 2003
|
|
1IVZ
 
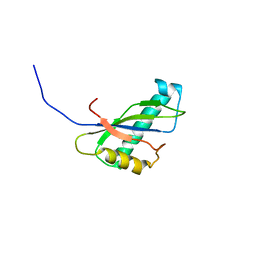 | | Solution structure of the SEA domain from murine hypothetical protein homologous to human mucin 16 | | Descriptor: | hypothetical protein 1110008I14RIK | | Authors: | Maeda, T, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-02 | | Release date: | 2002-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16)
J.Biol.Chem., 279, 2004
|
|
5X4Z
 
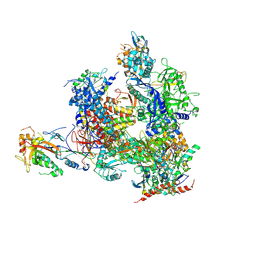 | | RNA Polymerase II from Komagataella Pastoris (Type-1 crystal) | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, RNA polymerase II subunit, ... | | Authors: | Ehara, H, Umehara, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (7.8 Å) | | Cite: | Crystal structure of RNA polymerase II from Komagataella pastoris
Biochem. Biophys. Res. Commun., 487, 2017
|
|
3VA2
 
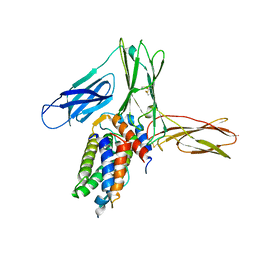 | | Crystal structure of human Interleukin-5 in complex with its alpha receptor | | Descriptor: | Interleukin-5, Interleukin-5 receptor subunit alpha | | Authors: | Kusano, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-12-28 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural basis of interleukin-5 dimer recognition by its alpha receptor
Protein Sci., 21, 2012
|
|
5X50
 
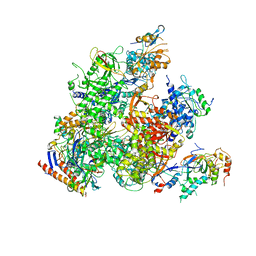 | | RNA Polymerase II from Komagataella Pastoris (Type-2 crystal) | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, RNA polymerase II subunit, ... | | Authors: | Ehara, H, Umehara, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.293 Å) | | Cite: | Crystal structure of RNA polymerase II from Komagataella pastoris
Biochem. Biophys. Res. Commun., 487, 2017
|
|
5X51
 
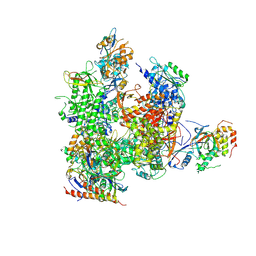 | | RNA Polymerase II from Komagataella Pastoris (Type-3 crystal) | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, RNA polymerase II subunit, ... | | Authors: | Ehara, H, Umehara, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6.996 Å) | | Cite: | Crystal structure of RNA polymerase II from Komagataella pastoris
Biochem. Biophys. Res. Commun., 487, 2017
|
|
5X60
 
 | | Crystal structure of LSD1-CoREST in complex with peptide 9 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2017-02-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
1WV9
 
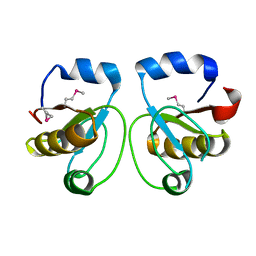 | | Crystal Structure of Rhodanese Homolog TT1651 from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 | | Descriptor: | Rhodanese Homolog TT1651 | | Authors: | Mizohata, E, Hattori, M, Tatsuguchi, A, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-12-12 | | Release date: | 2005-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the single-domain rhodanese homologue TTHA0613 from Thermus thermophilus HB8
Proteins, 64, 2006
|
|
1WGU
 
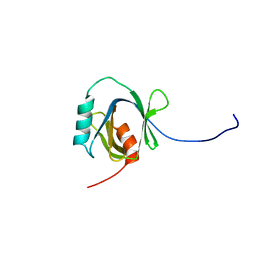 | | Solution Structure of the C-terminal Phosphotyrosine Interaction Domain of APBB2 from Mouse | | Descriptor: | amyloid beta (A4) precursor protein-binding, family B, member 2 | | Authors: | Li, H, Hayashi, F, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
1WNA
 
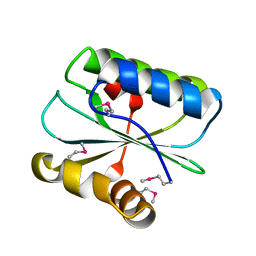 | | Crystal structure of the hypothetical protein TT1805 from Thermus thermophillus HB8 | | Descriptor: | the hypothetical protein (TT1805) | | Authors: | Pioszak, A.A, Kishishita, S, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-28 | | Release date: | 2005-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the hypothetical protein TT1805 from Thermus thermophillus HB8
To be Published
|
|
2KBO
 
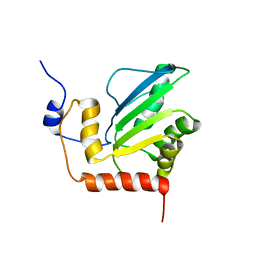 | | Structure, interaction, and real-time monitoring of the enzymatic reaction of wild type APOBEC3G | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Furukawa, A, Nagata, T, Matsugami, A, Habu, Y, Sugiyama, R, Hayashi, F, Kobayashi, N, Yokoyama, S, Takaku, H, Katahira, M. | | Deposit date: | 2008-12-04 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G
Embo J., 28, 2009
|
|
1X56
 
 | |
1WFQ
 
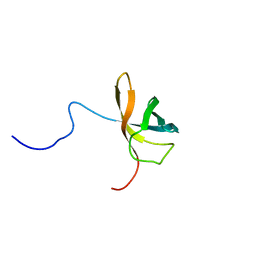 | | Solution structure of the first cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | UNR protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
1WJX
 
 | | Crystal sturucture of TT0801 from Thermus thermophilus | | Descriptor: | POTASSIUM ION, SsrA-binding protein | | Authors: | Bessho, Y, Shibata, R, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1WOT
 
 | | Structure of putative minimal nucleotidyltransferase | | Descriptor: | PUTATIVE MINIMAL NUCLEOTIDYLTRANSFERASE | | Authors: | Suzuki, S, Hatanaka, H, Hondoh, T, Okumura, A, Kuroda, Y, Kuramitsu, S, Shibata, T, Inoue, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-08-25 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of Putative Minimal Nucleotidyltransferase
To be Published
|
|
1WWY
 
 | | Solution structure of the DUF1000 domain of a thioredoxin-like protein 1 | | Descriptor: | Thioredoxin-like protein 1 | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal DUF1000 domain of the human thioredoxin-like 1 protein.
Proteins, 78, 2010
|
|
