2H9H
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | Hepatitis A virus protease 3C, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Three residue peptide | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-06-09 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
J.Mol.Biol., 361, 2006
|
|
2H6M
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | N-ACETYL-LEUCYL-ALANYL-ALANYL-(N,N-DIMETHYL)-GLUTAMINE-(1,4-DIOXO-3,4-DIHYDRO-1H-PHTHALAZIN-2-YL)METHYLKETONE INHIBITOR, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Picornain 3C | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-05-31 | | Release date: | 2006-08-08 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors.
J.Mol.Biol., 361, 2006
|
|
1TJN
 
 | | Crystal structure of hypothetical protein af0721 from Archaeoglobus fulgidus | | Descriptor: | Sirohydrochlorin cobaltochelatase | | Authors: | Yin, J, Xu, X.L, Cuff, M, Walker, J.R, Edwards, A, Savchenko, A, James, M.N.G, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-06 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of af0721: a hypothetical protein bearing sequence similarity with class II chelatases in cobalamin synthesis
To be Published
|
|
3FHA
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, GLYCEROL, ... | | Authors: | Yin, J, Li, L, Shaw, N, Li, Y, Song, J.K, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.C, Wang, L.X, Wang, P, Liu, Z.J. | | Deposit date: | 2008-12-09 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A.
Plos One, 4, 2009
|
|
7SR8
 
 | | Molecular mechanism of the the wake-promoting agent TAK-925 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Hypocretin receptor type 2, ... | | Authors: | Yin, J, Chapman, K, Lian, P, De Brabander, J.K, Rosenbaum, D.M. | | Deposit date: | 2021-11-08 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
1NGX
 
 | | Chimeric Germline Fab 7g12 with jeffamine fragment bound | | Descriptor: | Germline Metal Chelatase Catalytic Antibody, Heavy chain, Light chain, ... | | Authors: | Yin, J, Andryski, S.E, Beuscher, A.B, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2002-12-18 | | Release date: | 2003-03-18 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for substrate strain in antibody catalysis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NGY
 
 | | Chimeric Mature Fab 7g12-Apo | | Descriptor: | Mature Metal Chelatase Catalytic Antibody, Heavy chain, Light chain | | Authors: | Yin, J, Andryski, S.A, Beuscher, A.B, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2002-12-18 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural evidence for substrate strain in antibody catalysis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1N7M
 
 | | Germline 7G12 with N-methylmesoporphyrin | | Descriptor: | Germline Metal Chelatase Catalytic Antibody, chain H, chain L, ... | | Authors: | Yin, J, Andryski, S.E, Beuscher IV, A.E, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2002-11-15 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for substrate strain in antibody catalysis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NGZ
 
 | | Chimeric Germline Fab 7g12-apo | | Descriptor: | Germline Metal Chelatase Catalytic Antibody, Heavy chain, Light chain | | Authors: | Yin, J, Andryski, S.A, Beuscher, A.B, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2002-12-18 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural evidence for substrate strain in antibody catalysis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NGW
 
 | | Chimeric Affinity Matured Fab 7g12 complexed with mesoporphyrin | | Descriptor: | Mature Metal Chelatase Catalytic Antibody, Heavy chain, Light chain, ... | | Authors: | Yin, J, Andryski, S.E, Beuscher IV, A.E, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2002-12-18 | | Release date: | 2003-02-04 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural evidence for substrate strain in antibody catalysis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4QP1
 
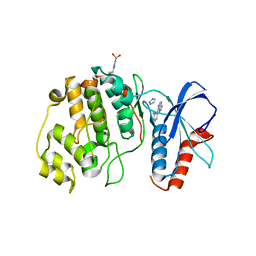 | |
4QP9
 
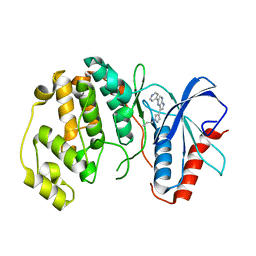 | |
4QP4
 
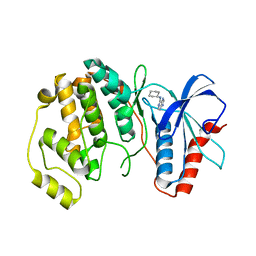 | |
4QPA
 
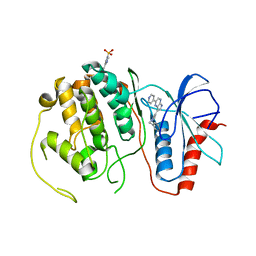 | |
4QP7
 
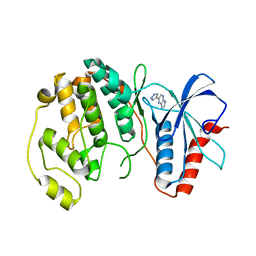 | |
4QP3
 
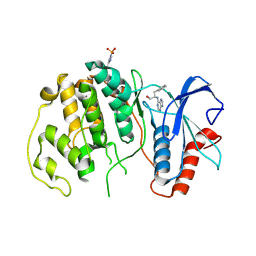 | |
4QP6
 
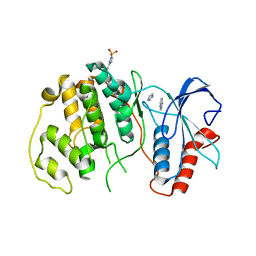 | |
4QP2
 
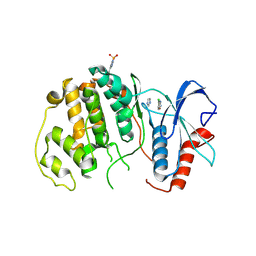 | |
4QP8
 
 | |
4O6E
 
 | | Discovery of 5,6,7,8-tetrahydropyrido[3,4-d]pyrimidine Inhibitors of Erk2 | | Descriptor: | Mitogen-activated protein kinase 1, N-[(1S)-1-(3-chloro-4-fluorophenyl)-2-hydroxyethyl]-2-(tetrahydro-2H-pyran-4-ylamino)-5,8-dihydropyrido[3,4-d]pyrimidine-7(6H)-carboxamide | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2013-12-20 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of 5,6,7,8-tetrahydropyrido[3,4-d]pyrimidine inhibitors of Erk2.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3TMO
 
 | | The catalytic domain of human deubiquitinase DUBA | | Descriptor: | OTU domain-containing protein 5 | | Authors: | Yin, J, Bosanac, I, Ma, X, Hymowitz, S, Starovasnik, M, Cochran, A. | | Deposit date: | 2011-08-31 | | Release date: | 2012-01-11 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phosphorylation-dependent activity of the deubiquitinase DUBA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4E55
 
 | |
4E57
 
 | |
4E56
 
 | |
8V52
 
 | |
