1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1NBS
 
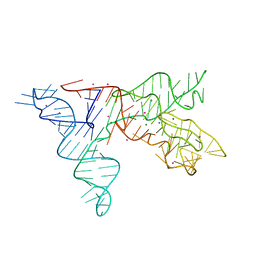 | | Crystal structure of the specificity domain of Ribonuclease P RNA | | Descriptor: | LEAD (II) ION, MAGNESIUM ION, RIBONUCLEASE P RNA | | Authors: | Krasilnikov, A.S, Yang, X, Pan, T, Mondragon, A. | | Deposit date: | 2002-12-03 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the specificity domain of Ribonuclease P
Nature, 421, 2003
|
|
5EB0
 
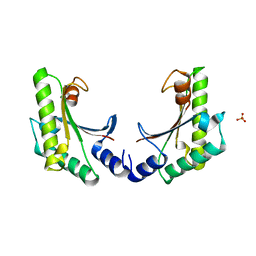 | | crystal form II of YfiB belonging to P41 | | Descriptor: | SULFATE ION, YfiB | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB2
 
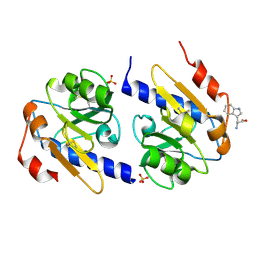 | | Trp-bound YfiR | | Descriptor: | SULFATE ION, TRYPTOPHAN, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.709 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
4ROU
 
 | |
4RYL
 
 | | Human Protein Arginine Methyltransferase 3 in complex with 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea | | Descriptor: | 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dobrovetsky, E, Kaniskan, H.U, Szewczyk, M, Yu, Z, Eram, M.S, Yang, X, Schmidt, K, Luo, X, Dai, M, He, F, Zang, I, Lin, Y, Kennedy, S, Li, F, Tempel, W, Smil, D, Min, S.J, Landon, M, Lin-Jones, J, Huang, X.P, Roth, B.L, Schapira, M, Atadja, P, Barsyte-Lovejoy, D, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Zhao, K, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent, Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 3 (PRMT3).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5EB1
 
 | | the YfiB-YfiR complex | | Descriptor: | SULFATE ION, YfiB, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB3
 
 | | VB6-bound protein | | Descriptor: | 4,5-bis(hydroxymethyl)-2-methyl-pyridin-3-ol, SULFATE ION, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5B6G
 
 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, GLYCEROL, PHQ-ALA-GLY-GLU-ALA-XYC-TYR-GLU, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
6LPI
 
 | | Crystal Structure of AHAS holo-enzyme | | Descriptor: | Acetolactate synthase isozyme 1 large subunit, Acetolactate synthase isozyme 1 small subunit, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zhang, Y, Yang, X, Xi, Z, Shen, Y. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Molecular architecture of the acetohydroxyacid synthase holoenzyme.
Biochem.J., 477, 2020
|
|
5IWZ
 
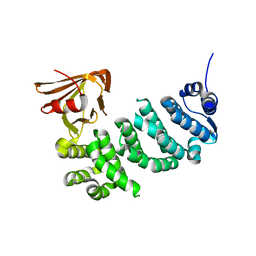 | | Synaptonemal complex protein | | Descriptor: | Synaptonemal complex protein 2 | | Authors: | Feng, J, Fu, S, Cao, X, Wu, H, Lu, J, Zeng, M, Liu, L, Yang, X, Shen, Y. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Structure of synaptonemal complexes protein at 2.6 angstroms resolution
To Be Published
|
|
5IZ8
 
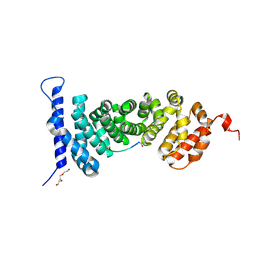 | | Protein-protein interaction | | Descriptor: | ACE-ALA-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein, TRIETHYLENE GLYCOL | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZ9
 
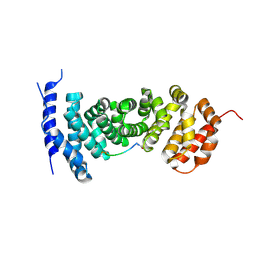 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZ6
 
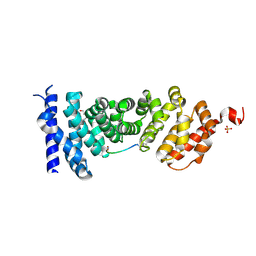 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, DI(HYDROXYETHYL)ETHER, PHQ-ALA-GLY-GLU-ALA-LEU-TYR-GLU-NH2, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5T4D
 
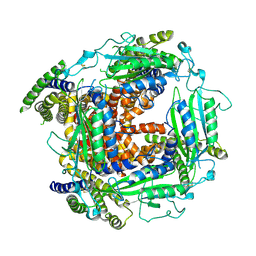 | | Cryo-EM structure of Polycystic Kidney Disease protein 2 (PKD2), residues 198-703 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hPKD:198-703, Polycystin-2 | | Authors: | Shen, P.S, Yang, X, DeCaen, P.G, Liu, X, Bulkley, D, Clapham, D.E, Cao, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs.
Cell, 167, 2016
|
|
5IZA
 
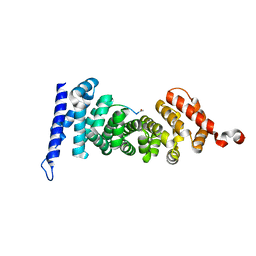 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-TRP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5J0A
 
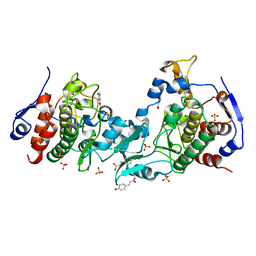 | | Crystal structure of PDZ-binding kinase | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Lymphokine-activated killer T-cell-originated protein kinase, SULFATE ION | | Authors: | Zou, Q.W, Zhou, H, Yang, X. | | Deposit date: | 2016-03-28 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The crystal structure of an inactive dimer of PDZ-binding kinase
Biochem.Biophys.Res.Commun., 476, 2016
|
|
1RXQ
 
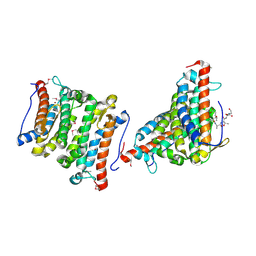 | | YfiT from Bacillus subtilis is a probable metal-dependent hydrolase with an unusual four-helix bundle topology | | Descriptor: | ALANINE, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Rajan, S.S, Yang, X, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-18 | | Release date: | 2004-07-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | YfiT from Bacillus subtilis Is a Probable Metal-Dependent Hydrolase with an Unusual Four-Helix Bundle Topology
Biochemistry, 43, 2004
|
|
1R5K
 
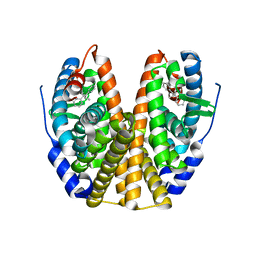 | | Human Estrogen Receptor alpha Ligand-Binding Domain In Complex With GW5638 | | Descriptor: | (2E)-3-{4-[(1E)-1,2-DIPHENYLBUT-1-ENYL]PHENYL}ACRYLIC ACID, Estrogen receptor | | Authors: | Wu, Y.-L, Yang, X, Ren, Z, McDonnell, D.P, Norris, J.D, Willson, T.M, Greene, G.L. | | Deposit date: | 2003-10-10 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for an unexpected mode of SERM-mediated ER antagonism.
Mol.Cell, 18, 2005
|
|
1RKT
 
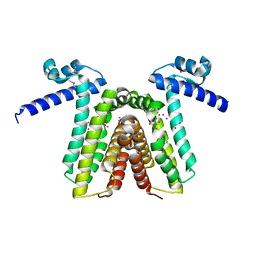 | | Crystal structure of yfiR, a putative transcriptional regulator from Bacillus subtilis | | Descriptor: | UNKNOWN ATOM OR ION, protein yfiR | | Authors: | Anderson, W.F, Rajan, S.S, Yang, X, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-23 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of YfiR, an unusual TetR/CamR-type putative transcriptional regulator from Bacillus subtilis.
Proteins, 65, 2006
|
|
2ZKS
 
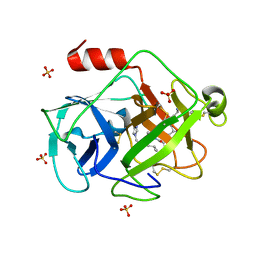 | | Structural insights into the proteolytic machinery of apoptosis-inducing Granzyme M | | Descriptor: | Granzyme M, SULFATE ION, hGzmM inhibitor | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Yang, X, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-03-28 | | Release date: | 2009-03-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
7C2M
 
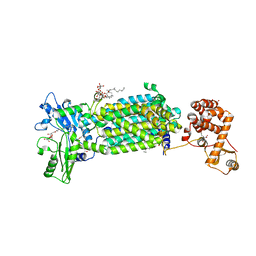 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with NITD-349 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Chimera of drug exporters of the RND superfamily-like protein and Endolysin, N-(4,4-dimethylcyclohexyl)-4,6-bis(fluoranyl)-1H-indole-2-carboxamide, ... | | Authors: | Zhang, B, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
7C2N
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SPIRO | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1'-(2,3-dihydro-1,4-benzodioxin-6-ylmethyl)spiro[6,7-dihydrothieno[3,2-c]pyran-4,4'-piperidine], Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
3BV6
 
 | | Crystal structure of uncharacterized metallo protein from Vibrio cholerae with beta-lactamase like fold | | Descriptor: | FE (III) ION, Metal-dependent hydrolase | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Yang, X, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of uncharacterized metallo protein from Vibrio cholerae with beta-lactamase like fold.
To be Published
|
|
1U8X
 
 | | CRYSTAL STRUCTURE OF GLVA FROM BACILLUS SUBTILIS, A METAL-REQUIRING, NAD-DEPENDENT 6-PHOSPHO-ALPHA-GLUCOSIDASE | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, Maltose-6'-phosphate glucosidase, ... | | Authors: | Rajan, S.S, Yang, X, Collart, F, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-09 | | Release date: | 2004-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Novel Catalytic Mechanism of Glycoside Hydrolysis Based on the Structure of an NAD(+)/Mn(2+)-Dependent Phospho-alpha-Glucosidase from Bacillus subtilis.
STRUCTURE, 12, 2004
|
|
