2NZ0
 
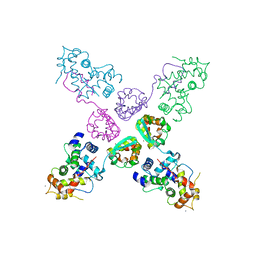 | | Crystal structure of potassium channel Kv4.3 in complex with its regulatory subunit KChIP1 | | Descriptor: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | Authors: | Wang, H, Yan, Y, Shen, Y, Chen, L, Wang, K. | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for modulation of Kv4 K(+) channels by auxiliary KChIP subunits.
Nat.Neurosci., 10, 2007
|
|
5YM0
 
 | | The crystal structure of DHAD | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, SULFATE ION | | Authors: | Zang, X, Huang, W.X, Cheng, R, Wu, L, Zhou, J.H, Tang, Y, Yan, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | The crystal structure of DHAD
To Be Published
|
|
8HS0
 
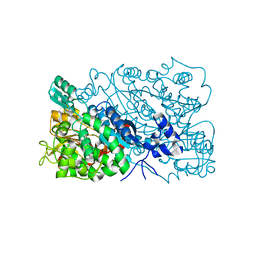 | | The mutant structure of DHAD V178W | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural bases of dihydroxy acid dehydratase inhibition and biodesign for self-resistance
Biodes Res, 0, 2024
|
|
8IKZ
 
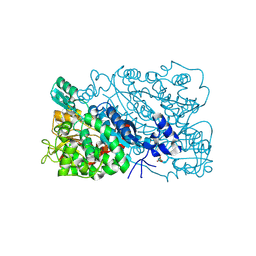 | | The mutant structure of DHAD | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural bases of dihydroxy acid dehydratase inhibition and biodesign for self-resistance
Biodes Res, 0, 2024
|
|
8IMU
 
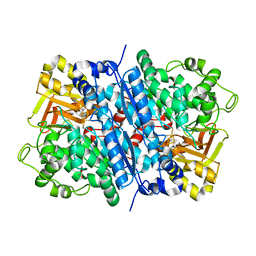 | | Dihydroxyacid dehydratase (DHAD) mutant-V497F | | Descriptor: | ACETATE ION, Dihydroxy-acid dehydratase, chloroplastic, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural bases of dihydroxy acid dehydratase inhibition and biodesign for self-resistance
Biodes Res, 0, 2024
|
|
1T28
 
 | | High resolution structure of a picornaviral internal cis-acting replication element | | Descriptor: | 34-MER | | Authors: | Thiviyanathan, V, Yang, Y, Kaluarachchi, K, Reynbrand, R, Gorenstein, D.G, Lemon, S.M. | | Deposit date: | 2004-04-20 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of a picornaviral internal cis-acting replication element(cre).
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1NI8
 
 | | H-NS dimerization motif | | Descriptor: | DNA-binding protein H-NS | | Authors: | Bloch, V, Yang, Y, Margeat, E, Chavanieu, A, Aug, M.T, Robert, B, Arold, S, Rimsky, S, Kochoyan, M. | | Deposit date: | 2002-12-22 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The H-NS dimerisation domain defines a new fold contributing to DNA recognition
Nat.Struct.Biol., 10, 2003
|
|
5WBT
 
 | |
2HKO
 
 | | Crystal structure of LSD1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1 | | Authors: | Chen, Y, Yang, Y.T, Wang, F, Yanane, K, Zhang, Y, Lei, M. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human histone lysine-specific demethylase 1 (LSD1).
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6LMS
 
 | |
5WTB
 
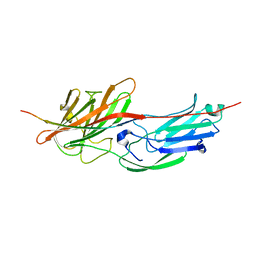 | | Complex Structure of Staphylococcus aureus SdrE with human complement factor H | | Descriptor: | Peptide from Complement factor H, Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
8J2K
 
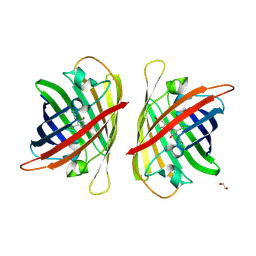 | | Crystal structure of a bright green fluorescent protein (StayGold) with double mutation (N137A, Q140S) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, StayGold(N137A, Q140S) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2I
 
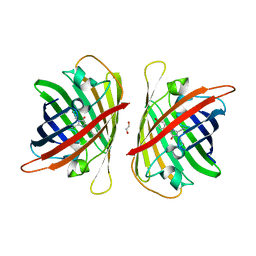 | | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (Q140S) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, StayGold(Q140S) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J3J
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with double mutations (Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, StayGold(Q140S, Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2L
 
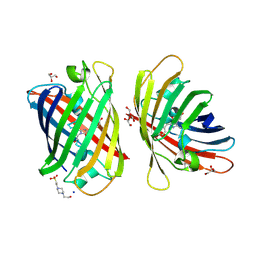 | | Crystal structure of a bright green fluorescent protein (StayGold) with double mutations (N137A, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2H
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (N137A) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | GLYCEROL, SODIUM ION, StayGold(N137A) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J86
 
 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*CP*TP*TP*AP*AP*TP*CP*TP*CP*AP*CP*AP*TP*AP*GP*CP*AP*GP*CP*TP*)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-04-30 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
8J8G
 
 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 in complex with a DNA duplex and cidofovir diphosphate | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*CP*TP*TP*AP*AP*TP*CP*TP*CP*AP*CP*AP*TP*AP*GP*CP*AP*GP*CP*T)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-05-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
8J8F
 
 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 in complex with a DNA duplex and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-05-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
7Y8L
 
 | | Structure of ScIRED-R2-V3 from Streptomyces clavuligerus in complex with 5-(2,5-difluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 5-[2,5-bis(fluoranyl)phenyl]-3,4-dihydro-2~{H}-pyrrole, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
7Y8M
 
 | | Structure of ScIRED-R2-V3 from Streptomyces clavuligerus in complex with 5-(3-fluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 2-[2,5-bis(fluoranyl)phenyl]pyrrolidine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, reductase | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
7Y40
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, staygold | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8JYE
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 and BTN2A1 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, Butyrophilin subfamily 2 member A1, ... | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.-C, Guo, R.-T, Zhang, Y.H. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYC
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 and BTN2A1 in Complex with DMAPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 2 member A1, Butyrophilin subfamily 3 member A1, ... | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.-C, Guo, R.-T, Zhang, Y.H. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
6XV2
 
 | | Full structure of RYMV P1 protein, derived from crystallographic and NMR data. | | Descriptor: | ZINC ION, p1 | | Authors: | Poignavent, V, Hoh, F, Vignols, F, Demene, H, Yang, Y, Gillet, F.X. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Flexible and Original Architecture of Two Unrelated Zinc Fingers Underlies the Role of the Multitask P1 in RYMV Spread.
J.Mol.Biol., 434, 2022
|
|
