6JHQ
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F4 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
8UA8
 
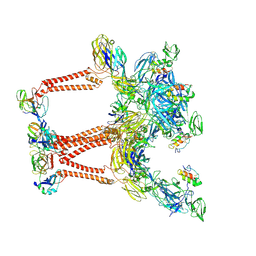 | | Structure of Semliki Forest virus VLP in complex with VLDLR LA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Assembly protein E3, ... | | Authors: | Abraham, J, Yang, P, Li, W, Fan, X, Pan, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of SFV VLP-VLDLRAD2 complex at the 3-fold axes
To Be Published
|
|
8UA4
 
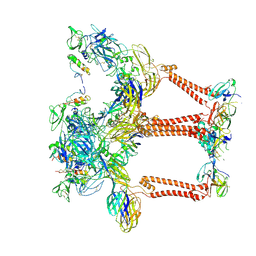 | | Structure of eastern equine encephalitis virus VLP in complex with VLDLR LA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Capsid protein, ... | | Authors: | Abraham, J, Yang, P, Li, W, Fan, X, Pan, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for VLDLR recognition by eastern equine encephalitis virus.
Nat Commun, 15, 2024
|
|
8UA9
 
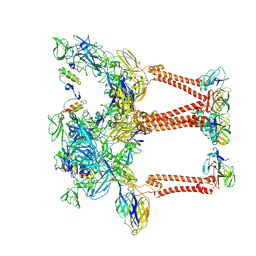 | | Structure of eastern equine encephalitis virus VLP unliganded quasi-threefold spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein, Envelope glycoprotein E1, ... | | Authors: | Abraham, J, Yang, P, Li, W, Fan, X, Pan, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for VLDLR recognition by eastern equine encephalitis virus.
Nat Commun, 15, 2024
|
|
7LC9
 
 | |
3ROV
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
7MP7
 
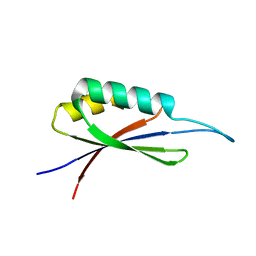 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sb3 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7MN1
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sa1 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7MQ4
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sb1 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
1DYR
 
 | | THE STRUCTURE OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE TO 1.9 ANGSTROMS RESOLUTION | | Descriptor: | DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIMETHOPRIM | | Authors: | Champness, J.N, Achari, A, Ballantine, S.P, Bryant, P.K, Delves, C.J, Stammers, D.K. | | Deposit date: | 1994-09-14 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of Pneumocystis carinii dihydrofolate reductase to 1.9 A resolution.
Structure, 2, 1994
|
|
6M5R
 
 | | The coordinates of the apo monomeric terminase complex | | Descriptor: | Tripartite terminase subunit 1, Tripartite terminase subunit 2, Tripartite terminase subunit 3, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
7MN2
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sb2 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
6M5T
 
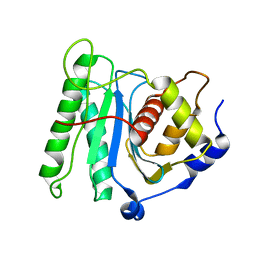 | | The coordinate of the nuclease domain of the apo terminase complex | | Descriptor: | Tripartite terminase subunit 3 | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M5U
 
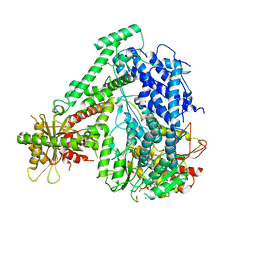 | | The coordinates of the monomeric terminase complex in the presence of the ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Zhu, L, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M5S
 
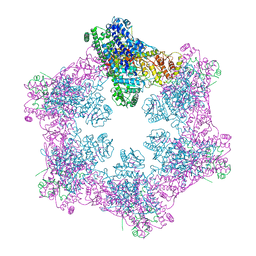 | | The coordinates of the apo hexameric terminase complex | | Descriptor: | Tripartite terminase subunit 1, Tripartite terminase subunit 2, Tripartite terminase subunit 3, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
1PGA
 
 | |
1PGB
 
 | |
6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6UAI
 
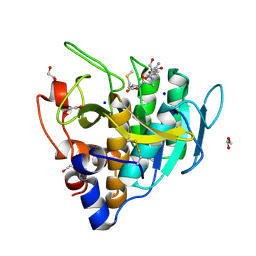 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with YSAM peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6UAO
 
 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with the peptide EEYSAM | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptide EEYSAM, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6UBE
 
 | | Azide-triggered subtilisin SUBT_BACAM complexed with the peptide LFRAL | | Descriptor: | AZIDE ION, GLYCEROL, Peptide LFRAL, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J, Gallagher, D.T, Custer, G. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6U9L
 
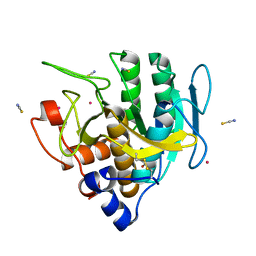 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM | | Descriptor: | GLYCEROL, POTASSIUM ION, SUBTILISIN BPN', ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
1DY3
 
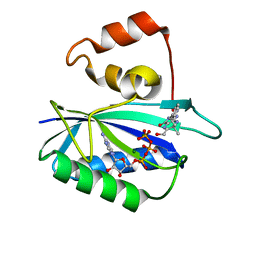 | | Ternary complex of 7,8-dihydro-6-hydroxymethylpterinpyrophosphokinase from Escherichia coli with ATP and a substrate analogue. | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 7,8-DIHYDRO-6-HYDROXYMETHYL-7-METHYL-7-[2-PHENYLETHYL]-PTERIN, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stammers, D.K, Achari, A, Somers, D.O, Bryant, P.K, Rosemond, J, Scott, D.L, Champness, J.N. | | Deposit date: | 2000-01-21 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A X-Ray Structure of the Ternary Complex of 7,8-Dihydro-6-Hydroxymethylpterinpyrophosphokinase from Escherichia Coli with ATP and a Substrate Analogue
FEBS Lett., 456, 1999
|
|
3P33
 
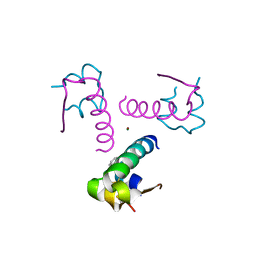 | | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Hua, Q.X, Wan, Z.L, Huang, K, Hu, S.Q, Phillip, N.F, Jia, W.H, Whittingham, J, Dodson, G.G, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2010-10-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity
To be Published
|
|
1SPB
 
 | | SUBTILISIN BPN' PROSEGMENT (77 RESIDUES) COMPLEXED WITH A MUTANT SUBTILISIN BPN' (266 RESIDUES). CRYSTAL PH 4.6. CRYSTALLIZATION TEMPERATURE 20 C DIFFRACTION TEMPERATURE-160 C | | Descriptor: | SODIUM ION, SUBTILISIN BPN', SUBTILISIN BPN' PROSEGMENT | | Authors: | Gallagher, D.T, Gilliland, G.L, Wang, L, Bryan, P.N. | | Deposit date: | 1995-06-21 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The prosegment-subtilisin BPN' complex: crystal structure of a specific 'foldase'.
Structure, 3, 1995
|
|
