2ZGU
 
 | |
2ZGM
 
 | | Crystal structure of recombinant Agrocybe aegerita lectin,rAAL, complex with lactose | | Descriptor: | Anti-tumor lectin, SULFATE ION, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Li, D.F, Yang, N, Wang, D.C. | | Deposit date: | 2008-01-23 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the tumor cell apoptosis-inducing activity of an antitumor lectin from the edible mushroom Agrocybe aegerita
J.Mol.Biol., 387, 2009
|
|
2ZGT
 
 | |
2ZGQ
 
 | |
2ZGO
 
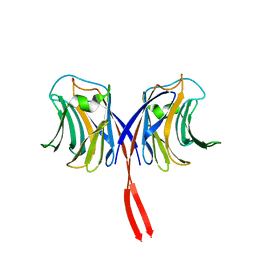 | | Crystal structure of AAL mutant H59Q complex with lactose | | Descriptor: | Anti-tumor lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Li, D.F, Yang, N, Wang, D.C. | | Deposit date: | 2008-01-23 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the tumor cell apoptosis-inducing activity of an antitumor lectin from the edible mushroom Agrocybe aegerita
J.Mol.Biol., 387, 2009
|
|
5B30
 
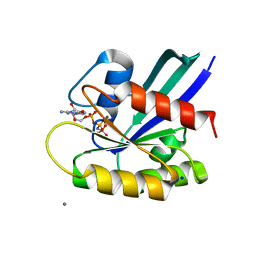 | | H-Ras WT in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Kumasaka, T, Miyano, N, Baba, S, Matsumoto, S, Kataoka, T, Shima, F. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Mechanism for Conformational Dynamics of Ras-GTP Elucidated from In-Situ Structural Transition in Crystal
Sci Rep, 6, 2016
|
|
5B2Z
 
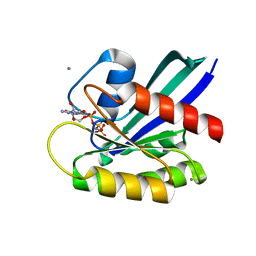 | | H-Ras WT in complex with GppNHp (state 2*) before structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Kumasaka, T, Miyano, N, Baba, S, Matsumoto, S, Kataoka, T, Shima, F. | | Deposit date: | 2016-02-07 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular Mechanism for Conformational Dynamics of Ras-GTP Elucidated from In-Situ Structural Transition in Crystal
Sci Rep, 6, 2016
|
|
4PW6
 
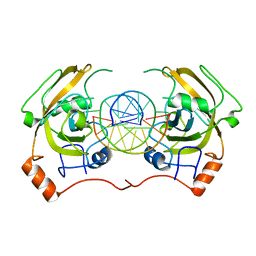 | | structure of UHRF2-SRA in complex with a 5hmC-containing DNA, complex II | | Descriptor: | 5hmC-containing DNA1, 5hmC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | Zhou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.789 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
4PW5
 
 | | structure of UHRF2-SRA in complex with a 5hmC-containing DNA, complex I | | Descriptor: | 5hmC-containing DNA1, 5hmC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | ZHou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
4PW7
 
 | | structure of UHRF2-SRA in complex with a 5mC-containing DNA | | Descriptor: | 5mC-containing DNA1, 5mC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | ZHou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-19 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
1KLS
 
 | | NMR Structure of the ZFY-6T[Y10L] Zinc Finger | | Descriptor: | ZINC FINGER Y-CHROMOSOMAL PROTEIN, ZINC ION | | Authors: | Lachenmann, M.J, Ladbury, J.E, Phillips, N.B, Narayana, N, Qian, X, Weiss, M.A. | | Deposit date: | 2001-12-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The hidden thermodynamics of a zinc finger.
J.Mol.Biol., 316, 2002
|
|
1KLR
 
 | | NMR Structure of the ZFY-6T[Y10F] Zinc Finger | | Descriptor: | ZINC FINGER Y-CHROMOSOMAL PROTEIN, ZINC ION | | Authors: | Lachenmann, M.J, Ladbury, J.E, Phillips, N.B, Narayana, N, Qian, X, Weiss, M.A. | | Deposit date: | 2001-12-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The hidden thermodynamics of a zinc finger.
J.Mol.Biol., 316, 2002
|
|
4Q5W
 
 | | Crystal structure of extended-Tudor 9 of Drosophila melanogaster | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Maternal protein tudor | | Authors: | Ren, R, Liu, H, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
4Q5Y
 
 | | Crystal structure of extended-Tudor 10-11 of Drosophila melanogaster | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H, Ren, R, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4IAO
 
 | | Crystal structure of Sir2 C543S mutant in complex with SID domain of Sir4 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent histone deacetylase SIR2, Regulatory protein SIR4, ... | | Authors: | Hsu, H.C, Wang, C.L, Wang, M, Yang, N, Chen, Z, Sternglanz, R, Xu, R.M. | | Deposit date: | 2012-12-07 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structural basis for allosteric stimulation of Sir2 activity by Sir4 binding
Genes Dev., 27, 2013
|
|
4HGA
 
 | | Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Liu, C.P, Xiong, C.Y, Wang, M.Z, Yu, Z.L, Yang, N, Chen, P, Zhang, Z.G, Li, G.H, Xu, R.M. | | Deposit date: | 2012-10-07 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1TD6
 
 | | Crystal structure of the conserved hypothetical protein MP506/MPN330 (gi: 1674200)from Mycoplasma pneumoniae | | Descriptor: | Hypothetical protein MG237 homolog | | Authors: | Das, D, Oganesyan, N, Yokota, H, Jancarik, J, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the conserved hypothetical protein MPN330 (GI: 1674200) from Mycoplasma pneumoniae.
Proteins, 58, 2004
|
|
1SU0
 
 | | Crystal structure of a hypothetical protein at 2.3 A resolution | | Descriptor: | NifU like protein IscU, ZINC ION | | Authors: | Liu, J, Oganesyan, N, Shin, D.-H, Jancarik, J, Pufan, R, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of an iron-sulfur cluster assembly protein IscU in a zinc-bound form.
Proteins, 59, 2005
|
|
5ZBA
 
 | | Crystal structure of Rtt109-Asf1-H3-H4-CoA complex | | Descriptor: | COENZYME A, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
5ZBB
 
 | | Crystal structure of Rtt109-Asf1-H3-H4 complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
5ZB9
 
 | | Crystal structure of Rtt109 from Aspergillus fumigatus | | Descriptor: | DNA damage response protein Rtt109, putative, GLYCEROL | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
1TPU
 
 | | S96P CHANGE IS A SECOND-SITE SUPPRESSOR FOR H95N SLUGGISH MUTANT TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the H95N lesion by the secondary S96P mutation in triosephosphate isomerase.
Biochemistry, 35, 1996
|
|
1TPV
 
 | | S96P CHANGE IS A SECOND-SITE SUPPRESSOR FOR H95N SLUGGISH MUTANT TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the H95N lesion by the secondary S96P mutation in triosephosphate isomerase.
Biochemistry, 35, 1996
|
|
1TPW
 
 | | TRIOSEPHOSPHATE ISOMERASE DRINKS WATER TO KEEP HEALTHY | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of water in the catalytic efficiency of triosephosphate isomerase.
Biochemistry, 38, 1999
|
|
