8IQU
 
 | | Structure of MtbFadD23 with PhU-AMS | | 分子名称: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, Fatty-acid-CoA ligase FadD23 | | 著者 | Yan, M.R, Zhang, W. | | 登録日 | 2023-03-17 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Structural basis for the development of potential inhibitors targeting FadD23 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
3QML
 
 | | The structural analysis of Sil1-Bip complex reveals the mechanism for Sil1 to function as a novel nucleotide exchange factor | | 分子名称: | 78 kDa glucose-regulated protein homolog, MAGNESIUM ION, Nucleotide exchange factor SIL1, ... | | 著者 | Yan, M, Li, J.Z, Sha, B.D. | | 登録日 | 2011-02-04 | | 公開日 | 2011-06-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor.
Biochem.J., 438, 2011
|
|
3QFP
 
 | |
3QFU
 
 | | Crystal structure of Yeast Hsp70 (Bip/kar2) complexed with ADP | | 分子名称: | 78 kDa glucose-regulated protein homolog, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Yan, M, Li, J.Z, Sha, B.D. | | 登録日 | 2011-01-22 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor.
Biochem.J., 438, 2011
|
|
8HCZ
 
 | |
8HDF
 
 | | Full length crystal structure of mycobacterium tuberculosis FadD23 in complex with ANP and PLM | | 分子名称: | Long-chain-fatty-acid--AMP ligase FadD23, PALMITIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | 登録日 | 2022-11-04 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
8HD4
 
 | | Full-length crystal structure of mycobacterium tuberculosis FadD23 in complex with AMPC16 | | 分子名称: | Long-chain-fatty-acid--AMP ligase FadD23, palmitoyl adenylate | | 著者 | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | 登録日 | 2022-11-03 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
8HEF
 
 | | The Crystal structure of deuterated S-217622 (Ensitrelvir) bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | 分子名称: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | 著者 | Yan, M, Zhang, H. | | 登録日 | 2022-11-08 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Synthesis of deuterated S-217622 (Ensitrelvir) with antiviral activity against coronaviruses including SARS-CoV-2.
Antiviral Res., 213, 2023
|
|
4PSX
 
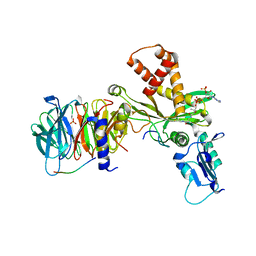 | | Crystal structure of histone acetyltransferase complex | | 分子名称: | COENZYME A, Histone H3, Histone H4, ... | | 著者 | Yang, M, Li, Y. | | 登録日 | 2014-03-08 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.509 Å) | | 主引用文献 | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
4PSW
 
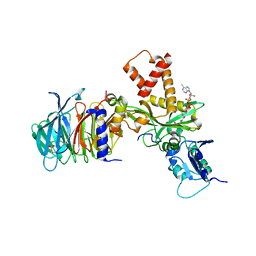 | | Crystal structure of histone acetyltransferase complex | | 分子名称: | COENZYME A, Histone H4 type VIII, Histone acetyltransferase type B catalytic subunit, ... | | 著者 | Yang, M, Li, Y. | | 登録日 | 2014-03-08 | | 公開日 | 2014-07-09 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
7ZNF
 
 | |
2RET
 
 | | The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the Type 2 Secretion System of Vibrio vulnificus | | 分子名称: | CHLORIDE ION, EpsJ, Pseudopilin EpsI, ... | | 著者 | Yanez, M.E, Korotkov, K.V, Abendroth, J, Hol, W.G.J. | | 登録日 | 2007-09-27 | | 公開日 | 2008-02-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus.
J.Mol.Biol., 375, 2008
|
|
5ILA
 
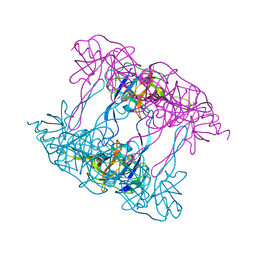 | | Deg9 protease domain | | 分子名称: | Protease Do-like 9 | | 著者 | Ouyang, M, Liu, L, Li, X.Y, Zhao, S, Zhang, L.X. | | 登録日 | 2016-03-04 | | 公開日 | 2017-03-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
6G32
 
 | | Crystal structure of human geranylgeranyl diphosphate synthase mutant D188Y | | 分子名称: | GLYCEROL, Geranylgeranyl pyrophosphate synthase | | 著者 | Lisnyansky, M, Kapelushnik, N, Ben-Bassat, A, Marom, M, Loewenstein, A, Khananshvili, D, Giladi, M, Haitin, Y. | | 登録日 | 2018-03-24 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.281 Å) | | 主引用文献 | Reduced Activity of Geranylgeranyl Diphosphate Synthase Mutant Is Involved in Bisphosphonate-Induced Atypical Fractures.
Mol. Pharmacol., 94, 2018
|
|
6G31
 
 | | Crystal structure of human geranylgeranyl diphosphate synthase mutant D188Y bound to zoledronate | | 分子名称: | Geranylgeranyl pyrophosphate synthase, MAGNESIUM ION, ZOLEDRONIC ACID | | 著者 | Lisnyansky, M, Kapelushnik, N, Ben-Bassat, A, Marom, M, Loewenstein, A, Khananshvili, D, Giladi, M, Haitin, Y. | | 登録日 | 2018-03-24 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Reduced Activity of Geranylgeranyl Diphosphate Synthase Mutant Is Involved in Bisphosphonate-Induced Atypical Fractures.
Mol. Pharmacol., 94, 2018
|
|
6PIK
 
 | | Tetrameric cryo-EM ArnA | | 分子名称: | Bifunctional polymyxin resistance protein ArnA, UDP-4-amino-4-deoxy-L-arabinose formyltransferase | | 著者 | Yang, M, Gehring, K. | | 登録日 | 2019-06-26 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
J.Struct.Biol., 208, 2019
|
|
6PIH
 
 | | Hexameric ArnA cryo-EM structure | | 分子名称: | Bifunctional polymyxin resistance protein ArnA, UDP-4-amino-4-deoxy-L-arabinose formyltransferase | | 著者 | Yang, M, Gehring, K. | | 登録日 | 2019-06-26 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
J.Struct.Biol., 208, 2019
|
|
1HV2
 
 | | SOLUTION STRUCTURE OF YEAST ELONGIN C IN COMPLEX WITH A VON HIPPEL-LINDAU PEPTIDE | | 分子名称: | ELONGIN C, VON HIPPEL-LINDAU DISEASE TUMOR SUPPRESSOR | | 著者 | Botuyan, M.V, Mer, G, Yi, G.-S, Koth, C.M, Case, D.A, Edwards, A.M, Chazin, W.J, Arrowsmith, C.H. | | 登録日 | 2001-01-05 | | 公開日 | 2001-09-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of yeast elongin C in complex with a von Hippel-Lindau peptide.
J.Mol.Biol., 312, 2001
|
|
8ANC
 
 | |
8ANB
 
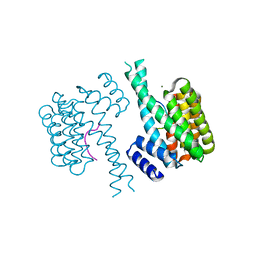 | |
2UXX
 
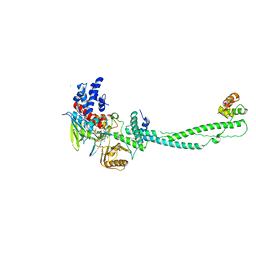 | | Human LSD1 Histone Demethylase-CoREST in complex with an FAD- tranylcypromine adduct | | 分子名称: | CHLORIDE ION, FAD-trans-2-Phenylcyclopropylamine Adduct, GLYCEROL, ... | | 著者 | Yang, M, Culhane, J.C, Machius, M, Cole, P.A, Yu, H. | | 登録日 | 2007-03-30 | | 公開日 | 2007-08-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Structural Basis for the Inhibition of the Lsd1 Histone Demethylase by the Antidepressant Trans-2-Phenylcyclopropylamine.
Biochemistry, 46, 2007
|
|
2UXN
 
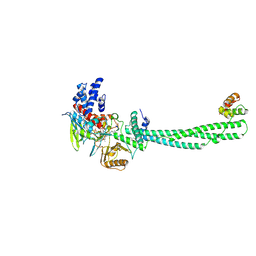 | | Structural Basis of Histone Demethylation by LSD1 Revealed by Suicide Inactivation | | 分子名称: | CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Yang, M, Culhane, J.C, Szewczuk, L.M, Gocke, C.B, Brautigam, C.A, Tomchick, D.R, Machius, M, Cole, P.A, Yu, H. | | 登録日 | 2007-03-28 | | 公開日 | 2007-05-29 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Structural Basis of Histone Demethylation by Lsd1 Revealed by Suicide Inactivation.
Nat.Struct.Mol.Biol., 14, 2007
|
|
7OJY
 
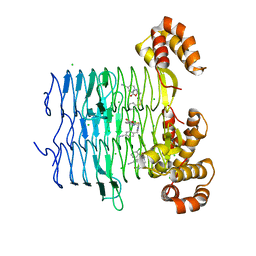 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 6 | | 分子名称: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-[(5-oxidanyl-1,3,4-oxadiazol-2-yl)methyl]ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION | | 著者 | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | 登録日 | 2021-05-17 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OK1
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 3 | | 分子名称: | Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION, ~{N}-[(5-azanyl-1,3,4-oxadiazol-2-yl)methyl]-2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]ethanamide | | 著者 | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | 登録日 | 2021-05-17 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OJ6
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 1 | | 分子名称: | 2-[2-(2-chlorophenyl)sulfanylethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-N-methyl-ethanamide, ACETATE ION, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, ... | | 著者 | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | 登録日 | 2021-05-14 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
