2ADZ
 
 | | solution structure of the joined PH domain of alpha1-syntrophin | | Descriptor: | Alpha-1-syntrophin | | Authors: | Yan, J, Wen, W, Xu, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
5IWW
 
 | | Crystal structure of RNA editing factor of designer PLS-type PPR/9R protein in complex with MORF9/RIP9 | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic, PLS9-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
5IWB
 
 | | Crystal structure of design pentatricopeptide repeat complex with MORF protein | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic, PLS3-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-22 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Structural basis for designer pentatricopeptide repeat proteins interaction with MORF protein
To be published
|
|
9J6I
 
 | | Crystal structure of the ABA receptor PYL1 in complex with DBSA compound | | Descriptor: | 5-bromanyl-N2-[(4-bromophenyl)-bis(oxidanyl)-$l^4-sulfanyl]-N1,N1,N3,N3-tetrakis(oxidanyl)benzene-1,2,3-triamine, Abscisic acid receptor PYL1 | | Authors: | Yan, J. | | Deposit date: | 2024-08-16 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stabilization of dimeric PYR/PYL/RCAR family members relieves abscisic acid-induced inhibition of seed germination.
Nat Commun, 15, 2024
|
|
5N41
 
 | | Archaeal DNA polymerase holoenzyme - SSO6202 at 1.35 Ang resolution | | Descriptor: | 1,2-ETHANEDIOL, PolB1 Binding Protein 2 | | Authors: | Yan, J, Beattie, T.R, Rojas, A.L, Schermerhorn, K, Gristwood, T, Trinidad, J.C, Albers, S.V, Roversi, P, Gardner, A.F, Abrescia, N.G.A, Bell, S.D. | | Deposit date: | 2017-02-09 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Identification and characterization of a heterotrimeric archaeal DNA polymerase holoenzyme.
Nat Commun, 8, 2017
|
|
5N35
 
 | | Gadolinium phased PBP2 (SSO6202) at 2.2 Ang | | Descriptor: | GADOLINIUM ATOM, GLYCEROL, NITRATE ION, ... | | Authors: | Yan, J, Beattie, T.R, Rojas, A.L, Schermerhorn, K, Gristwood, T, Trinidad, J.C, Albers, S.V, Roversi, P, Gardner, A.F, Abrescia, N.G.A, Bell, S.D. | | Deposit date: | 2017-02-08 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Identification and characterization of a heterotrimeric archaeal DNA polymerase holoenzyme.
Nat Commun, 8, 2017
|
|
5IZW
 
 | | Crystal structure of RNA editing specific factor of designer PLS-type PPR-9R protein | | Descriptor: | PLS9-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-26 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
3MGM
 
 | | Crystal structure of human NUDT16 | | Descriptor: | U8 snoRNA-decapping enzyme | | Authors: | Yan, J, Lu, G, Zhang, J, Qi, J, Li, Z, Gao, F. | | Deposit date: | 2010-04-07 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure and the template mRNA decapping activity of human NUDT16
To be Published
|
|
5GI0
 
 | | Crystal structure of RNA editing factor MORF9/RIP9 | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-06-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
2D2C
 
 | | Crystal Structure Of Cytochrome B6F Complex with DBMIB From M. Laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 2,5-DIBROMO-3-ISOPROPYL-6-METHYLBENZO-1,4-QUINONE, Apocytochrome f, ... | | Authors: | Yan, J, Kurisu, G, Cramer, W.A. | | Deposit date: | 2005-09-07 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Intraprotein transfer of the quinone analogue inhibitor 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone in the cytochrome b6f complex
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1Z87
 
 | | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | | Descriptor: | Alpha-1-syntrophin | | Authors: | Yan, J, Xu, W, Wen, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | Deposit date: | 2005-03-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
1Z86
 
 | | Solution structure of the PDZ domain of alpha-syntrophin | | Descriptor: | Alpha-1-syntrophin | | Authors: | Yan, J, Xu, W, Wen, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | Deposit date: | 2005-03-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
3KK9
 
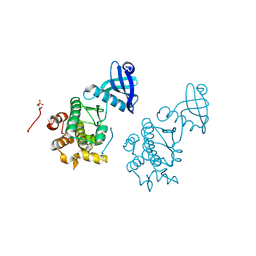 | | CaMKII Substrate Complex B | | Descriptor: | Calcium/calmodulin dependent protein kinase II | | Authors: | Kuriyan, J, Chao, L.H, Pellicena, P, Deindl, S, Barclay, L.A, Schulman, H. | | Deposit date: | 2009-11-04 | | Release date: | 2010-02-09 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6LPX
 
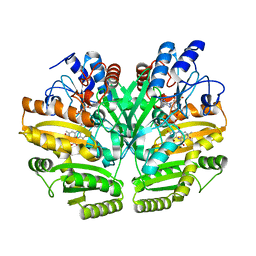 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with 2-oxoglutarate (2-OG) | | Descriptor: | 2-OXOGLUTARIC ACID, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPU
 
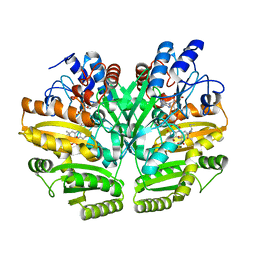 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with L-2-hydroxyglutarate (L-2-HG) | | Descriptor: | (2S)-2-HYDROXYPENTANEDIOIC ACID, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPP
 
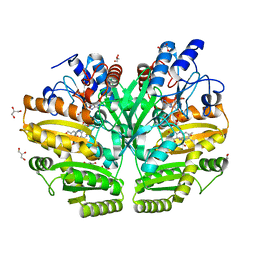 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-2-hydroxyglutarate (D-2-HG) | | Descriptor: | (2R)-2-hydroxypentanedioic acid, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPQ
 
 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-malate (D-MAL) | | Descriptor: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, D-MALATE, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPN
 
 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in apo form | | Descriptor: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPT
 
 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-lactate (D-LAC) | | Descriptor: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
8JXZ
 
 | | Chitin binding SusD-like protein AqSusD in complex with (GlcNAc)3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SusD-like protein AqSusD | | Authors: | Yang, J. | | Deposit date: | 2023-07-01 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights of a SusD-like protein in marine Bacteroidetes bacteria reveal the molecular basis for chitin recognition and acquisition.
Febs J., 291, 2024
|
|
8D9X
 
 | | Cryo-EM structure of human DELE1 in oligomeric form | | Descriptor: | Maltodextrin-binding protein,DAP3-binding cell death enhancer 1 short form | | Authors: | Yang, J, Lander, G.C. | | Deposit date: | 2022-06-11 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | DELE1 oligomerization promotes integrated stress response activation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8JDI
 
 | |
8JDH
 
 | |
8ZKC
 
 | | iron-sulfur cluster transfer protein ApbC | | Descriptor: | GLYCEROL, Iron-sulfur cluster carrier protein, MAGNESIUM ION | | Authors: | Yang, J, Liu, L. | | Deposit date: | 2024-05-16 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the iron-sulfur cluster transfer protein ApbC from Escherichia coli.
Biochem.Biophys.Res.Commun., 722, 2024
|
|
8JJC
 
 | | Tubulin-Y62 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(6,7-dimethoxy-3,4-dihydro-1~{H}-isoquinolin-2-yl)-6-(3-methoxyphenyl)pyrimidin-2-amine, CALCIUM ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-based design and synthesis of BML284 derivatives: A novel class of colchicine-site noncovalent tubulin degradation agents.
Eur.J.Med.Chem., 268, 2024
|
|
