4LN3
 
 | | The crystal structure of hemagglutinin from a H7N9 influenza virus (A/Shanghai/1/2013) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2013-07-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Analysis of the Hemagglutinin from the Recent 2013 H7N9 Influenza Virus.
J.Virol., 87, 2013
|
|
4LN4
 
 | | The crystal structure of hemagglutinin form a h7n9 influenza virus (a/shanghai/1/2013) in complex with lstb | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2013-07-11 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Analysis of the Hemagglutinin from the Recent 2013 H7N9 Influenza Virus.
J.Virol., 87, 2013
|
|
4M6W
 
 | | Crystal structure of the C-terminal segment of FANCM in complex with FAAP24 | | Descriptor: | Fanconi anemia group M protein, Fanconi anemia-associated protein of 24 kDa, SULFATE ION | | Authors: | Yang, H, Zhang, T, Tong, L, Ding, J. | | Deposit date: | 2013-08-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the functions of the FANCM-FAAP24 complex in DNA repair.
Nucleic Acids Res., 41, 2013
|
|
4GEZ
 
 | | Structure of a neuraminidase-like protein from A/bat/Guatemala/164/2009 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, H, Carney, P.J, Donis, R.O, Stevens, J. | | Deposit date: | 2012-08-02 | | Release date: | 2012-09-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4MC5
 
 | | Crystal structure of a subtype H18 hemagglutinin homologue from A/flat-faced bat/Peru/033/2010 (H18N11) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Stevens, J. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | New world bats harbor diverse influenza a viruses.
Plos Pathog., 9, 2013
|
|
4MC7
 
 | | Crystal structure of a subtype N11 neuraminidase-like protein of A/flat-faced bat/Peru/033/2010 (H18N11) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Stevens, J. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | New world bats harbor diverse influenza a viruses.
Plos Pathog., 9, 2013
|
|
4LXV
 
 | | Crystal Structure of the Hemagglutinin from a H1N1pdm A/WASHINGTON/5/2011 virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Chang, J.C, Guo, Z, Carney, P.J, Shore, D.A, Donis, R.O, Cox, N.J, Villanueva, J.M, Klimov, A.I, Stevens, J. | | Deposit date: | 2013-07-30 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Stability of Influenza A(H1N1)pdm09 Virus Hemagglutinins.
J.Virol., 88, 2014
|
|
2P9J
 
 | | Crystal structure of AQ2171 from Aquifex aeolicus | | Descriptor: | Hypothetical protein AQ2171 | | Authors: | Yang, H, Chen, L, Agari, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of AQ2171 from Aquifex aeolicus
To be Published
|
|
6K7C
 
 | | Dimeric Shewanella violacea cytochrome c5 | | Descriptor: | HEME C, NITRATE ION, Soluble cytochrome cA | | Authors: | Yang, H, Yamanaka, M, Nagao, S, Yasuhara, K, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2019-06-07 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Protein surface charge effect on 3D domain swapping in cells for c-type cytochromes.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
1WKC
 
 | | Crystal structure of a 5-formyltetrahydrofolate cycloligase-related protein from Thermus thermophilus HB8 | | Descriptor: | HB8 TT1367 protein, SULFATE ION | | Authors: | Yanai, H, Tsuge, H, Utsunomiya, H, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2004-12-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a 5-formyltetrahydrofolate cycloligase-related protein from Thermus thermophilus HB8
To be Published
|
|
7DL2
 
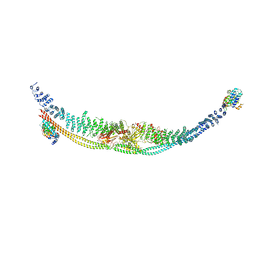 | | Cryo-EM structure of human TSC complex | | Descriptor: | Hamartin, Isoform 7 of Tuberin, TBC1 domain family member 7, ... | | Authors: | Yang, H, Yu, Z, Chen, X, Li, J, Li, N, Cheng, J, Gao, N, Yuan, H, Ye, D, Guan, K, Xu, Y. | | Deposit date: | 2020-11-25 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into TSC complex assembly and GAP activity on Rheb.
Nat Commun, 12, 2021
|
|
6ANW
 
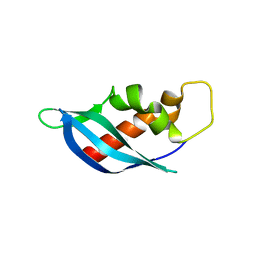 | | Crystal structure of anti-CRISPR protein AcrF10 | | Descriptor: | anti-CRISPR protein AcrF10 | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6ANV
 
 | | Crystal structure of anti-CRISPR protein AcrF1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
7D2T
 
 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, ... | | Authors: | Yang, H, Wei, Z, Yu, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2S
 
 | | Crystal structure of Rsu1/PINCH1_LIM5C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, Ras suppressor protein 1, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2U
 
 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, MALONATE ION, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
2CUW
 
 | | Crystal Structure of Thermus thermophilus PurS, one of the subunits of Formylglycinamide Ribonucleotide Amidotransferase in the purine biosynthetic pathway | | Descriptor: | PurS | | Authors: | Yanai, H, Kanagawa, M, Sampei, G, Kawai, G, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-30 | | Release date: | 2005-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Thermus thermophilus PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway
To be Published
|
|
2DGB
 
 | |
2CVK
 
 | |
7UU4
 
 | | Crystal structure of APOBEC3G complex with ssRNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*AP*AP*UP*UP*U)-3'), SULFATE ION, ... | | Authors: | Yang, H, Li, S, Chen, X.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
7UU3
 
 | | Crystal structure of APOBEC3G complex with 3'overhangs RNA-Complex | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(*CP*CP*CP*AP*CP*GP*GP*GP*AP*AP*U)-3'), RNA (5'-R(*CP*CP*CP*GP*UP*GP*GP*GP*AP*AP*U)-3'), ... | | Authors: | Yang, H, Li, S, Chen, X.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
7UU5
 
 | | Crystal structure of APOBEC3G complex with 5'-Overhang dsRNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*AP*AP*CP*CP*GP*CP*AP*GP*CP*G)-3'), RNA (5'-R(P*UP*AP*AP*CP*GP*CP*UP*GP*CP*GP*G)-3'), ... | | Authors: | Yang, H, Li, S, Chen, X.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | Descriptor: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | Deposit date: | 2020-03-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
1HKA
 
 | | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Descriptor: | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Authors: | Xiao, B, Shi, G, Chen, X, Yan, H, Ji, X. | | Deposit date: | 1998-09-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, a potential target for the development of novel antimicrobial agents.
Structure Fold.Des., 7, 1999
|
|
6UAL
 
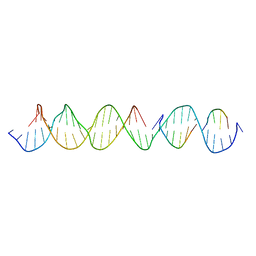 | | A Self-Assembling DNA Crystal Scaffold with Cavities Containing 3 Helical Turns per Edge | | Descriptor: | DNA (32-MER), DNA (5'-D(*TP*GP*GP*AP*AP*AP*CP*AP*GP*AP*CP*TP*GP*TP*CP*AP*GP*AP*TP*G)-3'), DNA (5'-D(P*AP*GP*CP*AP*TP*GP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4.514 Å) | | Cite: | Crystal Structure of a Self-Assembling DNA Crystal Scaffold with Rhombohedral Symmetry
To Be Published
|
|
