7JFW
 
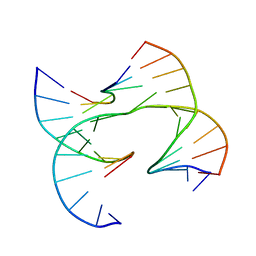 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J10 immobile Holliday junction | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*CP*AP*CP*TP*CP*A)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-17 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.012 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JH8
 
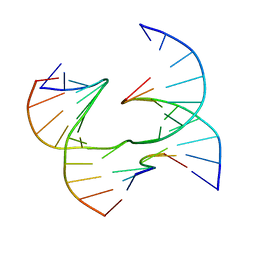 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J20 immobile Holliday junction | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*CP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*AP*GP*AP*CP*TP*CP*A)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-20 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.087 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JFU
 
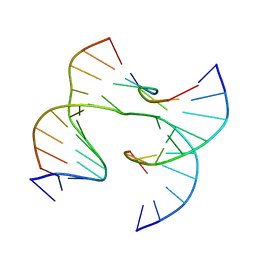 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J5 immobile Holliday junction | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*CP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*GP*GP*AP*CP*TP*CP*A)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-17 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.164 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JHB
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J24 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-20 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.114 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JFV
 
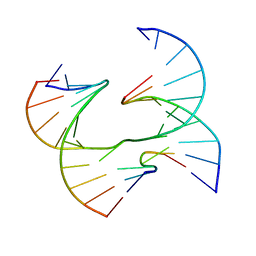 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J7 immobile Holliday junction | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*AP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*TP*CP*A)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-17 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JHC
 
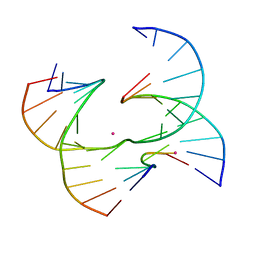 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J26 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*TP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*AP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-20 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JJZ
 
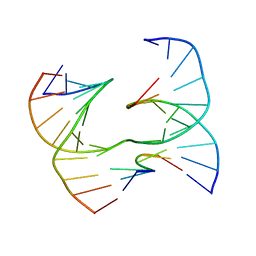 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J2 immobile Holliday junction | | 分子名称: | DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*CP*GP*TP*GP*TP*CP*GP*T)-3'), DNA (5'-D(P*CP*GP*AP*GP*GP*AP*CP*TP*C)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-27 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.068 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JFX
 
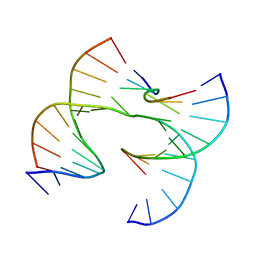 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J16 immobile Holliday junction | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*CP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*CP*CP*AP*CP*TP*CP*A)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-17 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JK0
 
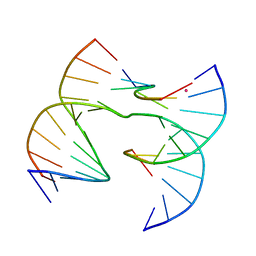 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J1 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*TP*GP*TP*GP*TP*CP*GP*T)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-27 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.058 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JFT
 
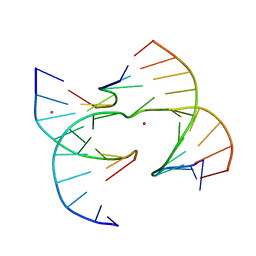 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J2 immobile Holliday junction | | 分子名称: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*CP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-17 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.158 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JH9
 
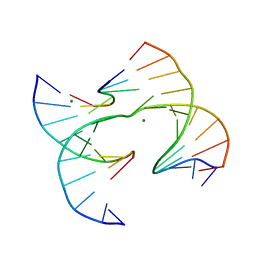 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J22 immobile Holliday junction | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*GP*CP*AP*CP*TP*CP*A)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2020-07-20 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.094 Å) | | 主引用文献 | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | 分子名称: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | 著者 | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | 登録日 | 2015-05-19 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.302 Å) | | 主引用文献 | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
1N3K
 
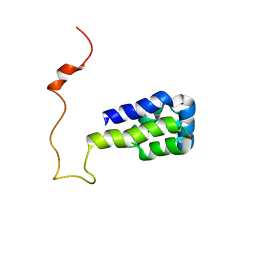 | | Solution structure of phosphoprotein enriched in astrocytes 15 kDa (PEA-15) | | 分子名称: | Astrocytic phosphoprotein PEA-15 | | 著者 | Hill, J.M, Vaidyanathan, H, Ramos, J.W, Ginsberg, M.H, Werner, M.H. | | 登録日 | 2002-10-28 | | 公開日 | 2003-01-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Recognition of ERK MAP Kinase by PEA-15 Reveals a Common Docking Site Within the Death Domain and Death Effector Domain
Embo J., 21, 2002
|
|
8ITU
 
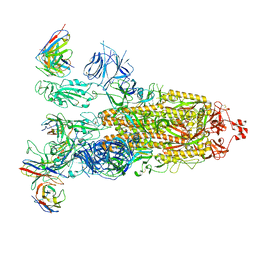 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 IgG. | | 分子名称: | 1H1 heavy chain, 1H1 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2023-03-23 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
6LU7
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | 登録日 | 2020-01-26 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
2RJF
 
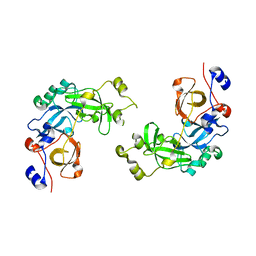 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 12-30), orthorhombic form I | | 分子名称: | Histone H4, Lethal(3)malignant brain tumor-like protein | | 著者 | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | 登録日 | 2007-10-14 | | 公開日 | 2007-10-30 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1VE6
 
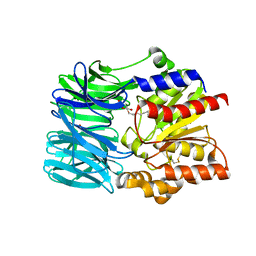 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 | | 分子名称: | Acylamino-acid-releasing enzyme, GLYCEROL, octyl beta-D-glucopyranoside | | 著者 | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | 登録日 | 2004-03-27 | | 公開日 | 2004-11-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
3CMT
 
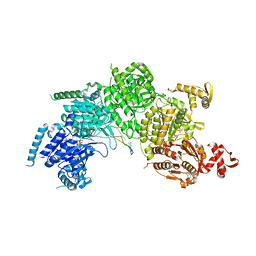 | | Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*DTP*DTP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DCP*DTP*DTP*DTP*DT)-3'), DNA (5'-D(P*DGP*DGP*DTP*DGP*DGP*DG)-3'), ... | | 著者 | Chen, Z, Yang, H, Pavletich, N.P. | | 登録日 | 2008-03-24 | | 公開日 | 2008-05-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures.
Nature, 453, 2008
|
|
5WBH
 
 | |
6M0K
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11b | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | 著者 | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | 登録日 | 2020-02-22 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.504 Å) | | 主引用文献 | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
2RJC
 
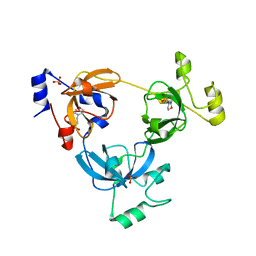 | | Crystal structure of L3MBTL1 protein in complex with MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | 著者 | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | 登録日 | 2007-10-14 | | 公開日 | 2007-10-30 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2RJD
 
 | | Crystal structure of L3MBTL1 protein | | 分子名称: | Lethal(3)malignant brain tumor-like protein | | 著者 | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | 登録日 | 2007-10-14 | | 公開日 | 2007-10-30 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2RUH
 
 | | Chemical Shift Assignments for MIP and MDM2 in bound state | | 分子名称: | E3 ubiquitin-protein ligase Mdm2 | | 著者 | Nagata, T, Shirakawa, K, Kobayashi, N, Shiheido, H, Horisawa, K, Katahira, M, Doi, N, Yanagawa, H. | | 登録日 | 2014-06-03 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for Inhibition of the MDM2:p53 Interaction by an Optimized MDM2-Binding Peptide Selected with mRNA Display
Plos One, 9, 2014
|
|
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | 分子名称: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | 著者 | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2020-10-04 | | 公開日 | 2021-04-21 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | 分子名称: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | 著者 | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2020-10-04 | | 公開日 | 2021-04-21 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
