6LMX
 
 | | Cryo-EM structure of the CALHM chimeric construct (9-mer) | | Descriptor: | Calcium homeostasis modulator 1,Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMV
 
 | | Cryo-EM structure of the C. elegans CLHM-1 | | Descriptor: | Calcium homeostasis modulator protein | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMW
 
 | | Cryo-EM structure of the CALHM chimeric construct (8-mer) | | Descriptor: | Calcium homeostasis modulator 1,Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMT
 
 | | Cryo-EM structure of the killifish CALHM1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Calcium homeostasis modulator 1 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
5Y78
 
 | | Crystal structure of the triose-phosphate/phosphate translocator in complex with inorganic phosphate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PHOSPHATE ION, Putative hexose phosphate translocator | | Authors: | Lee, Y, Nishizawa, T, Takemoto, M, Kumazaki, K, Yamashita, K, Hirata, K, Minoda, A, Nagatoishi, S, Tsumoto, K, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity
Nat Plants, 3, 2017
|
|
5YHA
 
 | | Crystal structure of Pd(allyl)/Wild Type Polyhedra | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PALLADIUM ION, ... | | Authors: | Abe, S, Atsumi, K, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of in cell protein crystals containing organometallic complexes.
Phys Chem Chem Phys, 20, 2018
|
|
5YRB
 
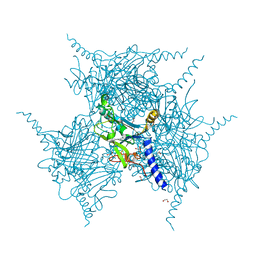 | | Crystal Structure of Oxidized Cypovirus Polyhedra R13A/E73C/Y83C/S193C/A194C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-02-21 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
5YRA
 
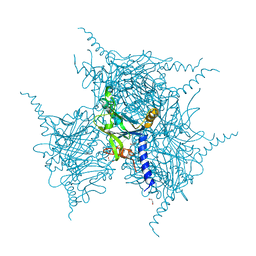 | | Crystal Structure of Cypovirus Polyhedra R13A/S193C/A194C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
5YRC
 
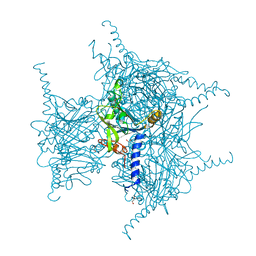 | | Crystal Structure of Oxidized Cypovirus Polyhedra R13A/E73C/Y83C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
5YR9
 
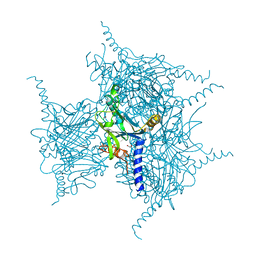 | | Crystal Structure of Cypovirus Polyhedra R13A/E73C/Y83C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-08 | | Release date: | 2018-02-21 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
5YHB
 
 | | Crystal structure of Pd(allyl)/polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | PALLADIUM ION, Palladium(II) allyl complex, Polyhedrin | | Authors: | Abe, S, Atsumi, K, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of in cell protein crystals containing organometallic complexes.
Phys Chem Chem Phys, 20, 2018
|
|
5YRD
 
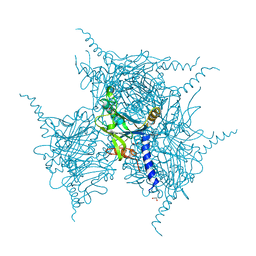 | | Crystal Structure of Oxidized Cypovirus Polyhedra R13A/S193C/A194C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
5YR1
 
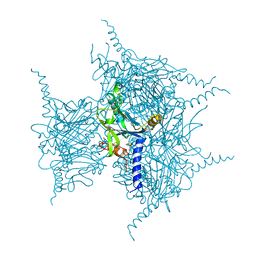 | | Crystal Structure of Cypovirus Polyhedra R13A/E73C/Y83C/S193C/A194C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-08 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
8C2I
 
 | | Transmembrane domain of resting state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C2H
 
 | | Transmembrane domain of active state homomeric GluA1 AMPA receptor in tandem with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C1P
 
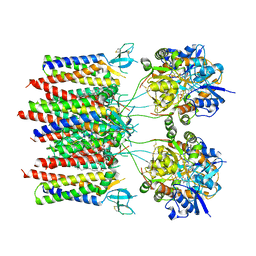 | | Active state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CYCLOTHIAZIDE, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C1S
 
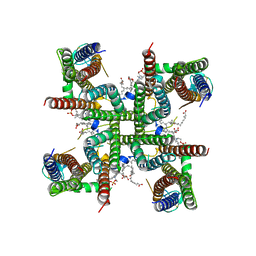 | | Transmembrane domain of resting state homomeric GluA2 F231A mutant AMPA receptor in complex with TARP gamma 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C1R
 
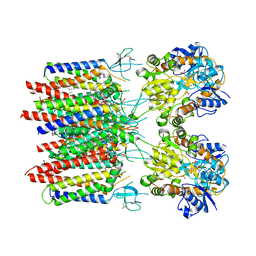 | | Resting state homomeric GluA2 F231A mutant AMPA receptor in complex with TARP gamma-2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C1Q
 
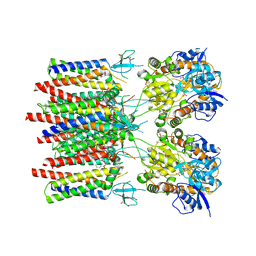 | | Resting state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
5B4X
 
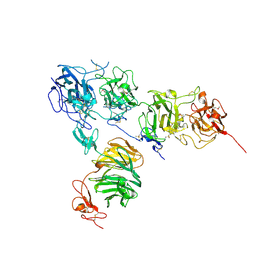 | | Crystal structure of the ApoER2 ectodomain in complex with the Reelin R56 fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Low density lipoprotein receptor-related protein 8, ... | | Authors: | Yasui, N, Hirai, H, Yamashita, K, Takagi, J, Nogi, T. | | Deposit date: | 2016-04-20 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the ectodomain from a LDLR close homologue in complex with its physiological ligand.
To Be Published
|
|
8W7N
 
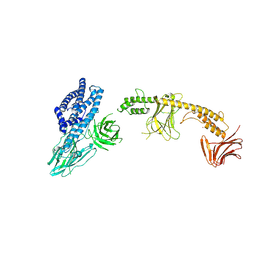 | | Crystal structure of the in-cell Cry1Aa purified from Bacillus thuringiensis | | Descriptor: | Pesticidal crystal protein Cry1Aa, UNKNOWN ATOM OR ION | | Authors: | Tanaka, J, Abe, S, Hayakawa, T, Kojima, M, Yamashita, K, Hirata, K, Ueno, T. | | Deposit date: | 2023-08-31 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the in-cell Cry1Aa purified from Bacillus thuringiensis.
Biochem.Biophys.Res.Commun., 685, 2023
|
|
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8IU0
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Nakamura, S, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2023-03-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
7VPK
 
 | | Cryo-EM structure of the human ATP13A2 (SPM-bound E2P state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
