3AP5
 
 | |
3AP9
 
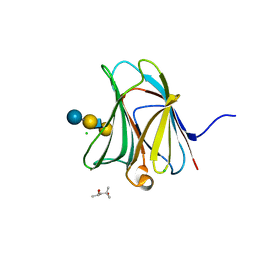 | | Crystal structure of the galectin-8 N-terminal carbohydrate recognition domain in complex with Lacto-N-fucopentaose III | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Galectin-8, ... | | Authors: | Matsuzaka, T, Ideo, H, Yamashita, K, Nonaka, T. | | Deposit date: | 2010-10-12 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Galectin-8-N-domain recognition mechanism for sialylated and sulfated glycans
J.Biol.Chem., 286, 2011
|
|
3A6J
 
 | | E122Q mutant creatininase complexed with creatine | | Descriptor: | Creatinine amidohydrolase, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6D
 
 | | Creatininase complexed with 1-methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6E
 
 | | W174F mutant creatininase, type I | | Descriptor: | CACODYLATE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6L
 
 | | E122Q mutant creatininase, Zn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, ZINC ION | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6H
 
 | | W154A mutant creatininase | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6K
 
 | | The E122Q mutant creatininase, Mn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6F
 
 | | W174F mutant creatininase, Type II | | Descriptor: | CACODYLATE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6G
 
 | | W154F mutant creatininase | | Descriptor: | Creatinine amidohydrolase, MANGANESE (II) ION, ZINC ION | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3APB
 
 | |
3B0V
 
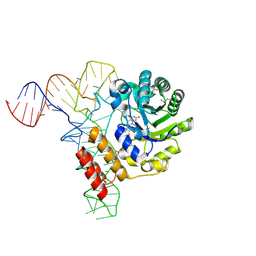 | | tRNA-dihydrouridine synthase from Thermus thermophilus in complex with tRNA | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA, tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-14 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B0U
 
 | | tRNA-dihydrouridine synthase from Thermus thermophilus in complex with tRNA fragment | | Descriptor: | FLAVIN MONONUCLEOTIDE, RNA (5'-R(*GP*GP*(H2U)P*A)-3'), tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-14 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B0P
 
 | | tRNA-dihydrouridine synthase from Thermus thermophilus | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-12 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8OFY
 
 | |
8OFX
 
 | | Molecular Mechanism of trypanosomal AQP2 | | Descriptor: | Aquaglyceroporin 2, [(2~{R},4~{S})-2-[4-[[4,6-bis(azanyl)-1,3,5-triazin-2-yl]amino]phenyl]-1,3,2-dithiarsolan-4-yl]methanol | | Authors: | Weyand, S.N, Matusevicius, M, Yamashita, K. | | Deposit date: | 2023-03-17 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular Mechanism of trypanosomal AQP2
To Be Published
|
|
8OFZ
 
 | |
7F47
 
 | | Cryo-EM structure of Rhizobium etli MprF | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Hypothetical conserved protein, [(2R)-1-[[(2R)-3-[(2S)-2,6-bis(azanyl)hexanoyl]oxy-2-oxidanyl-propoxy]-oxidanyl-phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (E)-octadec-9-enoate | | Authors: | Nishimura, M, Hirano, H, Kobayashi, K, Gill, C.P, Phan, C.N.K, Kise, Y, Kusakizako, T, Yamashita, K, Ito, Y, Roy, H, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-06-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of Rhizobium etli MprF
To Be Published
|
|
7EFC
 
 | | 1.70 A cryo-EM structure of streptavidin | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Kotecha, A, Nureki, O. | | Deposit date: | 2021-03-21 | | Release date: | 2021-04-28 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | 1.70 A cryo-EM structure of streptavidin using all frames (corresponding to 70 e/A^2 total dose)
To Be Published
|
|
7EFD
 
 | | 1.77 A cryo-EM structure of Streptavidin using first 40 frames (corresponding to about 40 e/A^2 total dose) | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Kotecha, A, Nureki, O. | | Deposit date: | 2021-03-21 | | Release date: | 2021-04-28 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.77 Å) | | Cite: | 1.77 A cryo-EM structure of Streptavidin using first 40 frames (corresponding to about 40e/A^2 total dose)
To Be Published
|
|
7DY0
 
 | |
7V6B
 
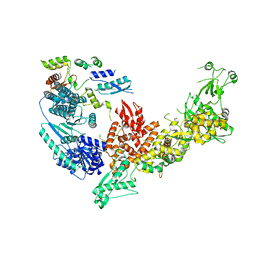 | | Structure of the Dicer-2-R2D2 heterodimer | | Descriptor: | Dicer-2, isoform A, R2D2 | | Authors: | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
7V6C
 
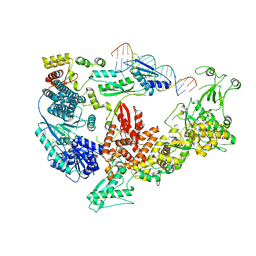 | | Structure of the Dicer-2-R2D2 heterodimer bound to small RNA duplex | | Descriptor: | Dicer-2, isoform A, R2D2, ... | | Authors: | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
