5XGR
 
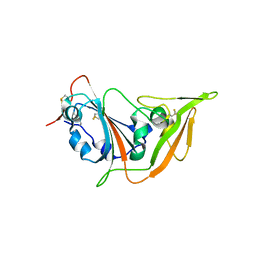 | | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Xue, H, Qi, J, Song, H, Qihui, W, Shi, Y, Gao, G.F. | | Deposit date: | 2017-04-16 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein
Virology, 507, 2017
|
|
8KFL
 
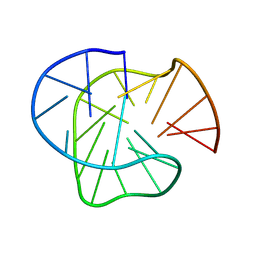 | | Stable G-quadruplex Conformation Formed in Promoter Region of Oncogene RET in 100 mM Na+ | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*AP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*G)-3') | | Authors: | Zhang, Y.P, Lan, W.X, Yin, S.W, Liu, Z.J, Xue, H.J, Cao, C.Y. | | Deposit date: | 2023-08-15 | | Release date: | 2024-09-04 | | Method: | SOLUTION NMR | | Cite: | Stable G-quadruplex Conformation Formed in Promoter Region of Oncogene RET Indicates New Target for Anti-cancer Drug Screening
To Be Published
|
|
8KF9
 
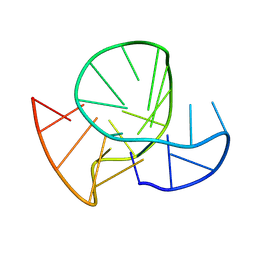 | | Stable G-quadruplex Conformation Formed in Promoter Region of Oncogene RET in 50 mM K+ solution | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*AP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*G)-3') | | Authors: | Zhang, Y.P, Lan, W.X, Yin, S.W, Liu, Z.J, Xue, H.J, Cao, C.Y. | | Deposit date: | 2023-08-15 | | Release date: | 2024-09-04 | | Method: | SOLUTION NMR | | Cite: | Stable G-quadruplex Conformation Formed in Promoter Region of Oncogene RET Indicates New Target for Anti-cancer Drug Screening
To Be Published
|
|
7X5C
 
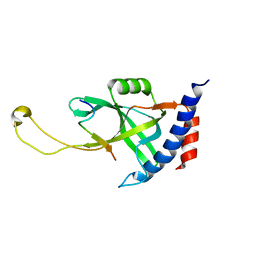 | | Solution structure of Tetrahymena p75OB1-p50PBM | | Descriptor: | Telomerase associated protein p50PBM, Telomerase-associated protein p75OB1 | | Authors: | Wu, B, Tang, T, Xue, H.J, Wu, J, Lei, M. | | Deposit date: | 2022-03-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Association of the CST complex and p50 in Tetrahymena is crucial for telomere maintenance
Structure, 2022
|
|
1NH9
 
 | | Crystal Structure of a DNA Binding Protein Mja10b from the hyperthermophile Methanococcus jannaschii | | Descriptor: | DNA-binding protein Alba | | Authors: | Wang, G, Bartlam, M, Guo, R, Yang, H, Xue, H, Liu, Y, Huang, L, Rao, Z. | | Deposit date: | 2002-12-19 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a DNA binding protein from the hyperthermophilic euryarchaeon Methanococcus jannaschii
Protein Sci., 12, 2003
|
|
3WFV
 
 | | HIV-1 CRF07 gp41 | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Du, J, Xue, H, Ma, J, Liu, F, Zhou, J, Shao, Y, Qiao, W, Liu, X. | | Deposit date: | 2013-07-24 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of HIV CRF07 B'/C gp41 reveals a hyper-mutant site in the middle of HR2 heptad repeat
Virology, 446, 2013
|
|
7ETT
 
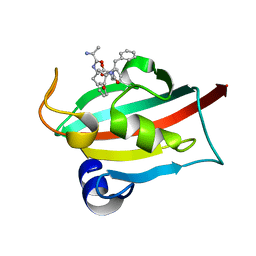 | | The FK1 domain of FKBP51 in complex with peptide-inhibitor hit QFPFV | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, peptide-inhibitor hit | | Authors: | Han, J.T, Zhu, Y.C, Pan, D.B, Xue, H.X, Wang, S, Liu, H.X, He, Y.X, Yao, X.J. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of pentapeptide-inhibitor hits targeting FKBP51 by combining computational modeling and X-ray crystallography.
Comput Struct Biotechnol J, 19, 2021
|
|
7ETV
 
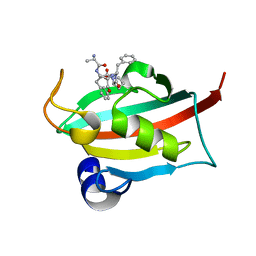 | | The FK1 domain of FKBP51 in complex with peptide-inhibitor hit DFPFV | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, peptide-inhibitor hit | | Authors: | Han, J.T, Zhu, Y.C, Pan, D.B, Xue, H.X, Wang, S, Liu, H.X, He, Y.X, Yao, X.J. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Discovery of pentapeptide-inhibitor hits targeting FKBP51 by combining computational modeling and X-ray crystallography.
Comput Struct Biotechnol J, 19, 2021
|
|
7ETU
 
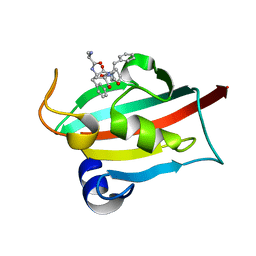 | | The FK1 domain of FKBP51 in complex with peptide-inhibitor hit SFPFT | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, peptide-inhibitor hit | | Authors: | Han, J.T, Zhu, Y.C, Pan, D.B, Xue, H.X, Wang, S, Liu, H.X, He, Y.X, Yao, X.J. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery of pentapeptide-inhibitor hits targeting FKBP51 by combining computational modeling and X-ray crystallography.
Comput Struct Biotechnol J, 19, 2021
|
|
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
6KIX
 
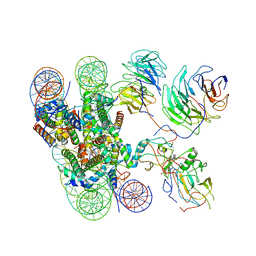 | | Cryo-EM structure of human MLL1-NCP complex, binding mode1 | | Descriptor: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIZ
 
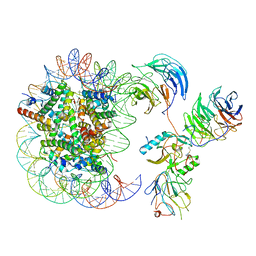 | | Cryo-EM structure of human MLL1-NCP complex, binding mode2 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIW
 
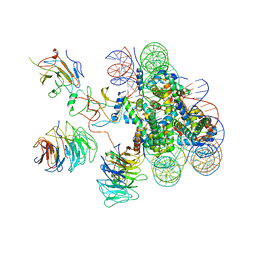 | | Cryo-EM structure of human MLL3-ubNCP complex (4.0 angstrom) | | Descriptor: | DNA (144-MER), DNA (145-MER), Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIV
 
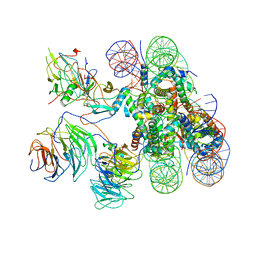 | | Cryo-EM structure of human MLL1-ubNCP complex (4.0 angstrom) | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIU
 
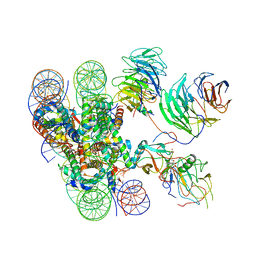 | | Cryo-EM structure of human MLL1-ubNCP complex (3.2 angstrom) | | Descriptor: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
