4NIQ
 
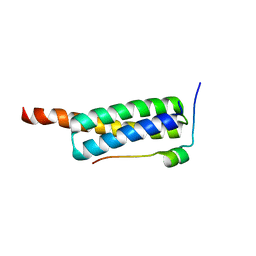 | | Crystal Structure of Vps4 MIT-Vfa1 MIM2 | | Descriptor: | VPS4-associated protein 1, Vacuolar protein sorting-associated protein 4 | | Authors: | Vild, C.J, Xu, Z. | | Deposit date: | 2013-11-06 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Vfa1 Binds to the N-terminal Microtubule-interacting and Trafficking (MIT) Domain of Vps4 and Stimulates Its ATPase Activity.
J.Biol.Chem., 289, 2014
|
|
7EO2
 
 | | Cryo-EM of Sphingosine 1-phosphate receptor 1 / Gi complex bound to FTY720p | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xu, Z, Ikuta, T. | | Deposit date: | 2021-04-21 | | Release date: | 2022-01-05 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural basis of sphingosine-1-phosphate receptor 1 activation and biased agonism.
Nat.Chem.Biol., 18, 2022
|
|
7EO4
 
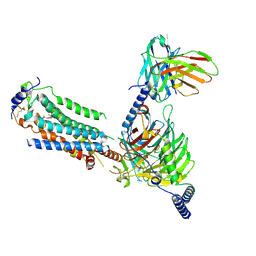 | | Cryo-EM of Sphingosine 1-phosphate receptor 1 / Gi complex bound to BAF312 | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xu, Z, Ikuta, T, Inoue, A. | | Deposit date: | 2021-04-21 | | Release date: | 2022-01-05 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of sphingosine-1-phosphate receptor 1 activation and biased agonism.
Nat.Chem.Biol., 18, 2022
|
|
4GVD
 
 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ in complex with Syndecan1 Peptide | | Descriptor: | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONIC ACID(DANSYL ACID), CHLORIDE ION, SODIUM ION, ... | | Authors: | Liu, X, Shepherd, T.R, Murray, A.M, Xu, Z, Fuentes, E.J. | | Deposit date: | 2012-08-30 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the Tiam1 PDZ domain/ phospho-syndecan1 complex reveals a ligand conformation that modulates protein dynamics.
Structure, 21, 2013
|
|
4GVC
 
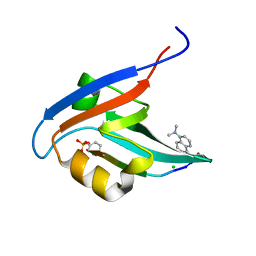 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ in complex with phosphorylated Syndecan1 Peptide | | Descriptor: | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONIC ACID(DANSYL ACID), CHLORIDE ION, SODIUM ION, ... | | Authors: | Liu, X, Shepherd, T.R, Murray, A.M, Xu, Z, Fuentes, E.J. | | Deposit date: | 2012-08-30 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The structure of the Tiam1 PDZ domain/ phospho-syndecan1 complex reveals a ligand conformation that modulates protein dynamics.
Structure, 21, 2013
|
|
5XS2
 
 | |
8JY0
 
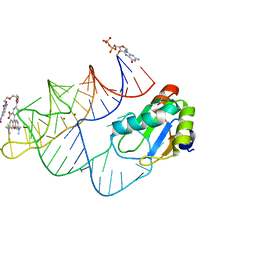 | | Crystal structure of RhoBAST complexed with TMR-DN | | Descriptor: | 2,4-dinitroaniline, 5-aminocarbonyl-2-[3-(dimethylamino)-6-dimethylazaniumylidene-xanthen-9-yl]benzoate, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Y, Xiao, Y, Xu, Z, Fang, X. | | Deposit date: | 2023-07-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanisms for binding and activation of a contact-quenched fluorophore by RhoBAST.
Nat Commun, 15, 2024
|
|
8HJE
 
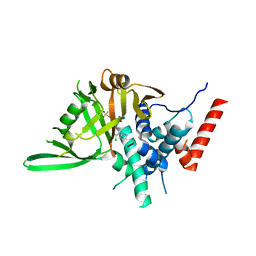 | | Vismodegib binds to the catalytical domain of human Ubiquitin-Specific Protease 28 | | Descriptor: | 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Shi, L, Wang, H, Xu, Z, Xiong, B, Zhang, N. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based discovery of potent USP28 inhibitors derived from Vismodegib.
Eur.J.Med.Chem., 254, 2023
|
|
5US1
 
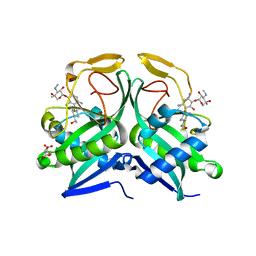 | | Crystal structure of aminoglycoside acetyltransferase AAC(2')-Ia in complex with N2'-acetylgentamicin C1A and coenzyme A | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-(acetylamino)-6-amino-2,3,4,6-tetradeoxy-alpha-D-erythro-hexopyranoside, ACETYL COENZYME *A, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Xu, Z, Wawrzak, Z, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Plazomicin Retains Antibiotic Activity against Most Aminoglycoside Modifying Enzymes.
ACS Infect Dis, 4, 2018
|
|
5YOC
 
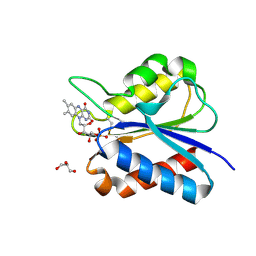 | | Crystal Structure of flavodoxin with engineered disulfide bond C102-R125C | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | Deposit date: | 2017-10-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
5YOG
 
 | | Crystal Structure of flavodoxin with engineered disulfide bond N14C-C93 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL, ... | | Authors: | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
5YO4
 
 | |
5YO5
 
 | |
5YOE
 
 | | Crystal Structure of flavodoxin with engineered disulfide bond A43C-L74C | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | Deposit date: | 2017-10-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
5YM7
 
 | |
5YO6
 
 | |
5YO3
 
 | | Crystal Structure of B562RIL with engineered disulfide bond V16C-A29C | | Descriptor: | SULFATE ION, Soluble cytochrome b562 | | Authors: | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
5YOB
 
 | | Crystal Structure of flavodoxin without engineered disulfide bond | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | Deposit date: | 2017-10-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.142 Å) | | Cite: | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
7XYQ
 
 | | Crystal strucutre of PD-L1 and the computationally designed DBL1_03 protein binder | | Descriptor: | ARGININE, CD274 molecule, DBL1_03 | | Authors: | Liu, K, Xu, Z, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S. | | Deposit date: | 2022-06-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7WF7
 
 | | Cryo-EM of Sphingosine 1-phosphate receptor 1 / Gi complex bound to S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xu, Z. | | Deposit date: | 2021-12-26 | | Release date: | 2022-01-05 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of sphingosine-1-phosphate receptor 1 activation and biased agonism.
Nat.Chem.Biol., 18, 2022
|
|
4YFJ
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib | | Descriptor: | Aminoglycoside 3'-N-acetyltransferase, SULFATE ION | | Authors: | Stogios, P.J, Xu, Z, Evdokimova, E, Yim, V, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib
To Be Published
|
|
6MN1
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA
To Be Published
|
|
6MN2
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with sisomicin-CoA | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with sisomicin-CoA
To Be Published
|
|
