8QN0
 
 | | Soluble epoxide hydrolase in complex with RK3 | | Descriptor: | (3~{a}~{R},6~{a}~{S})-~{N}-[(2,4-dichlorophenyl)methyl]-5-(4-methylphenyl)sulfonyl-1,3,3~{a},4,6,6~{a}-hexahydropyrrolo[3,4-c]pyrrole-2-carboxamide, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | Authors: | Kumar, A, Zhu, F, Ehrler, J.M.H, Li, F, Empel, C, Xu, Y, Atodiresei, I, Koenigs, R.M, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-25 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Photosensitization enables Pauson-Khand-type reactions with nitrenes.
Science, 383, 2024
|
|
8Y33
 
 | | A near-infrared fluorescent protein of de novo backbone design | | Descriptor: | 3-[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, near-infrared fluorescent protein | | Authors: | Hu, X, Xu, Y. | | Deposit date: | 2024-01-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Using Protein Design and Directed Evolution to Monomerize a Bright Near-Infrared Fluorescent Protein.
Acs Synth Biol, 13, 2024
|
|
8WTF
 
 | | The Crystal Structure of IRAK4 from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 1-{[(2S,3S,4S)-3-ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2023-10-18 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of IRAK4 from Biortus
To Be Published
|
|
8YGY
 
 | | Structure of the KLK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Kallikrein-1, SULFATE ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of the KLK1 from Biortus.
To Be Published
|
|
8YHF
 
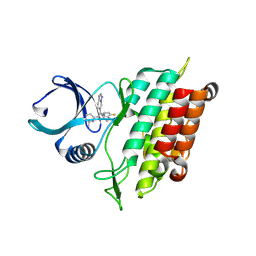 | | The Crystal Structure of the Type I TGF beta receptor from Biortus. | | Descriptor: | 2-fluoranyl-~{N}-[[5-(6-methylpyridin-2-yl)-4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-1~{H}-imidazol-2-yl]methyl]aniline, TGF-beta receptor type-1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Type I TGF beta receptor from Biortus.
To Be Published
|
|
8YHL
 
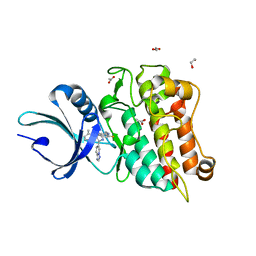 | | The Crystal Structure of Tgf-Beta Type I Receptor (Alk5) from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 2-fluoranyl-~{N}-[[5-(6-methylpyridin-2-yl)-4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-1~{H}-imidazol-2-yl]methyl]aniline, ACETATE ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of Tgf-Beta Type I Receptor (Alk5) from Biortus
To Be Published
|
|
8YHW
 
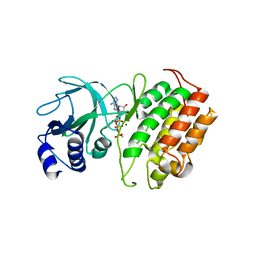 | | The Crystal Structure of NF-kB-inducing Kinase (NIK) from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, MAGNESIUM ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of NF-kB-inducing Kinase (NIK) from Biortus
To Be Published
|
|
8X87
 
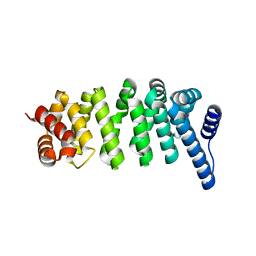 | |
8X88
 
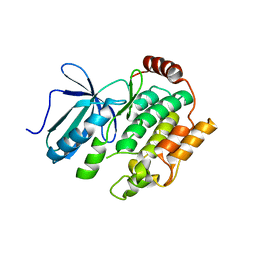 | |
8XPV
 
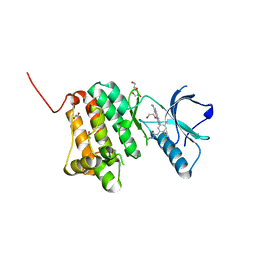 | | The Crystal Structure of EphA2 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 1-(3,3-dimethylbutyl)-3-{2-fluoro-4-methyl-5-[7-methyl-2-(methylamino)pyrido[2,3-d]pyrimidin-6-yl]phenyl}urea, Ephrin type-A receptor 2, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-01-04 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of EphA2 from Biortus.
To Be Published
|
|
1VRY
 
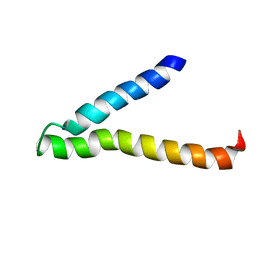 | | Second and Third Transmembrane Domains of the Alpha-1 Subunit of Human Glycine Receptor | | Descriptor: | Glycine receptor alpha-1 chain | | Authors: | Ma, D, Liu, Z, Li, L, Tang, P, Xu, Y. | | Deposit date: | 2005-07-20 | | Release date: | 2005-07-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Second and Third Transmembrane Domains of Human Glycine Receptor.
Biochemistry, 44, 2005
|
|
7BV1
 
 | | Cryo-EM structure of the apo nsp12-nsp7-nsp8 complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
7BV2
 
 | | The nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of Remdesivir(RTP) | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
4GU0
 
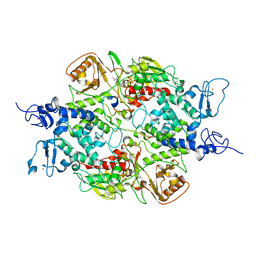 | | Crystal structure of LSD2 with H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Yang, H, Dong, Z, Fang, J, Zhu, T, Gong, W, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b
Cell Res., 23, 2013
|
|
4HSU
 
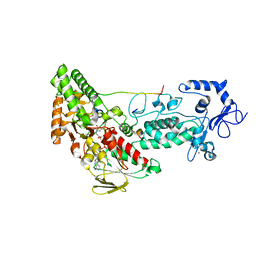 | | Crystal structure of LSD2-NPAC with H3(1-26)in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Xu, Y. | | Deposit date: | 2012-10-30 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b.
Cell Res., 23, 2013
|
|
4NDZ
 
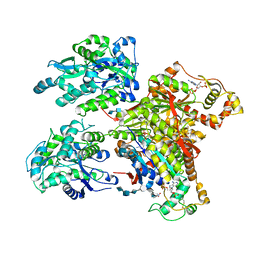 | | Structure of Maltose Binding Protein fusion to 2-O-Sulfotransferase with bound heptasaccharide and PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1 fusion, ... | | Authors: | Liu, C, Sheng, J, Krahn, J.M, Perera, L, Xu, Y, Hsieh, P, Liu, J, Pedersen, L.C. | | Deposit date: | 2013-10-28 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Deciphering the role of 2-O-sulfotransferase in regulating heparan sulfate biosynthesis
To be Published
|
|
4NM6
 
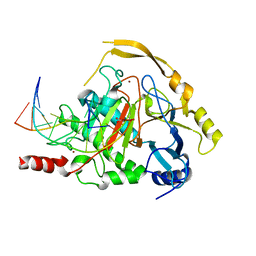 | | Crystal structure of TET2-DNA complex | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*(5CM)P*GP*GP*TP*GP*GP*T)-3', FE (II) ION, Methylcytosine dioxygenase TET2, ... | | Authors: | Hu, L, Li, Z, Cheng, J, Rao, Q, Gong, W, Liu, M, Wang, P, Xu, Y. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation.
Cell(Cambridge,Mass.), 155, 2013
|
|
4IA0
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-2-{2-ethoxy-5-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}-6-octylpyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ren, J, Chen, T, Xu, Y. | | Deposit date: | 2012-12-05 | | Release date: | 2014-01-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4I9Z
 
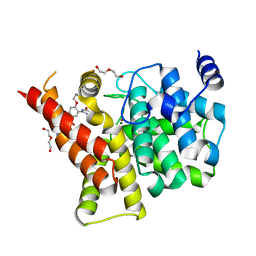 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-2-{5-[(4-methylpiperazin-1-yl)acetyl]-2-propoxyphenyl}-6-(propan-2-yl)pyrimidin-4(3H)-one, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Ren, J, Chen, T, Xu, Y. | | Deposit date: | 2012-12-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8AZ5
 
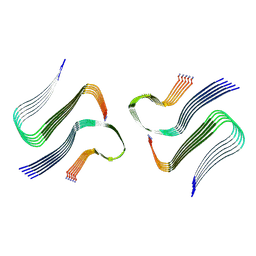 | | IAPP S20G plateau-phase fibril polymorph 4PF-CU | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
8AZ2
 
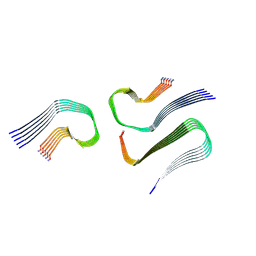 | | IAPP S20G growth-phase fibril polymorph 3PF-CU | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
8AWT
 
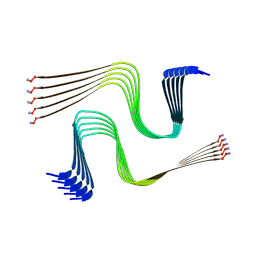 | | IAPP S20G lag-phase fibril polymorph 2PF-P | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-08-30 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
8AZ0
 
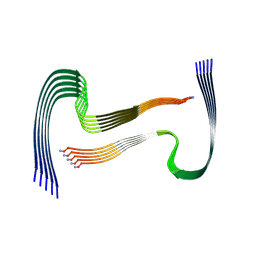 | | IAPP S20G growth-phase fibril polymorph 2PF-L | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
8AZ4
 
 | | IAPP S20G plateau-phase fibril polymorph 2PF-L | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
8AZ1
 
 | | IAPP S20G growth-phase fibril polymorph 2PF-C | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
