7DB7
 
 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase in complex with compound GDI05-001 | | Descriptor: | 1-[3-[2-(1H-indol-3-yl)ethylsulfamoyl]phenyl]-3-(1,3-thiazol-2-yl)urea, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, ... | | Authors: | Xu, M, Zhang, X, Xu, L, Chen, S. | | Deposit date: | 2020-10-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7DAW
 
 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase | | Descriptor: | Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, SULFATE ION | | Authors: | Xu, M, Zhang, X, Xu, L, Chen, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7DB8
 
 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase in complex with compound PF-3845 | | Descriptor: | N-pyridin-3-yl-4-[[3-[5-(trifluoromethyl)pyridin-2-yl]oxyphenyl]methyl]piperidine-1-carboxamide, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, ... | | Authors: | Xu, M, Zhang, X, Xu, L, Chen, S. | | Deposit date: | 2020-10-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
8I7U
 
 | |
6A8O
 
 | | Crystal structures of the serine protease domain of murine plasma kallikrein with peptide inhibitor mupain-1-16 | | Descriptor: | Plasma kallikrein, alpha-D-mannopyranose, peptide inhibitor,, ... | | Authors: | Xu, M, Jiang, L, Huang, M. | | Deposit date: | 2018-07-09 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of plasma kallikrein reveals the unusual flexibility of the S1 pocket triggered by Glu217.
Febs Lett., 592, 2018
|
|
8HEH
 
 | | Crystal structure of GCN5-related N-acetyltransferase 05790 | | Descriptor: | COENZYME A, GLYCEROL, GNAT family N-acetyltransferase | | Authors: | Xu, M.X, Ran, T.T, Wang, W. | | Deposit date: | 2022-11-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of prodigiosin binding protein PgbP, a GNAT family protein, in Serratia marcescens FS14.
Biochem.Biophys.Res.Commun., 640, 2022
|
|
8H0M
 
 | | Crystal structure of VioD | | Descriptor: | (2S)-2-ethylhexan-1-ol, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Xu, M, Ran, T, Wang, W. | | Deposit date: | 2022-09-29 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of the key enzyme VioD in the violacein synthesis pathway.
Proteins, 91, 2023
|
|
5GZS
 
 | | Structure of VC protein | | Descriptor: | ARGININE, GGDEF family protein | | Authors: | Xu, M, Wang, Y.Z, Yang, X.A, Xie, W, Jiang, T. | | Deposit date: | 2016-10-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural studies of the periplasmic portion of the diguanylate cyclase CdgH from Vibrio cholerae.
Sci Rep, 7, 2017
|
|
5EB3
 
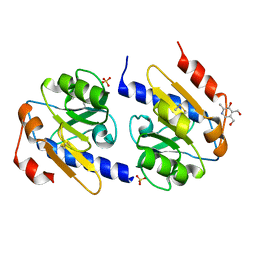 | | VB6-bound protein | | Descriptor: | 4,5-bis(hydroxymethyl)-2-methyl-pyridin-3-ol, SULFATE ION, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB0
 
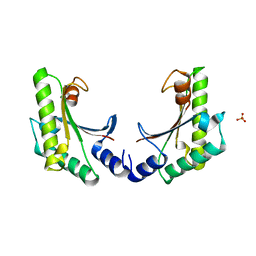 | | crystal form II of YfiB belonging to P41 | | Descriptor: | SULFATE ION, YfiB | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB2
 
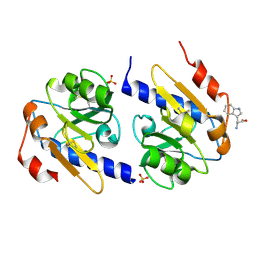 | | Trp-bound YfiR | | Descriptor: | SULFATE ION, TRYPTOPHAN, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.709 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB1
 
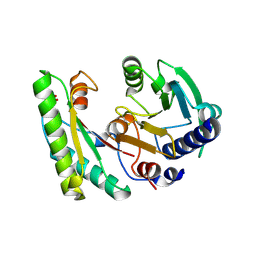 | | the YfiB-YfiR complex | | Descriptor: | SULFATE ION, YfiB, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EAZ
 
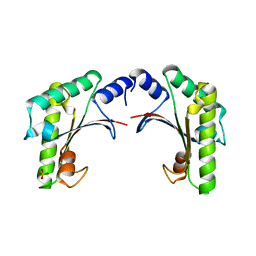 | | crystal form I of YfiB belonging to space groups P21 | | Descriptor: | SULFATE ION, YfiB | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
3GGR
 
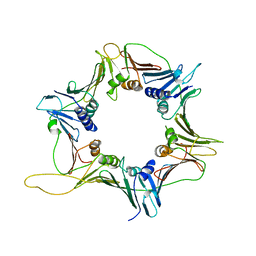 | | Crystal Structure of the Human Rad9-Hus1-Rad1 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1 | | Authors: | Xu, M, Bai, L, Hang, H.Y, Jiang, T. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and functional implications of the human rad9-hus1-rad1 cell cycle checkpoint complex
J.Biol.Chem., 284, 2009
|
|
7EL8
 
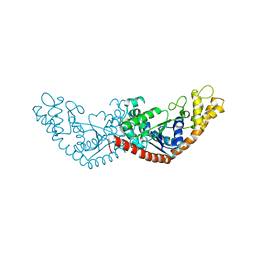 | |
7ENS
 
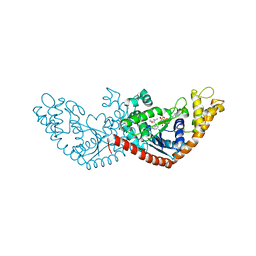 | |
7ELT
 
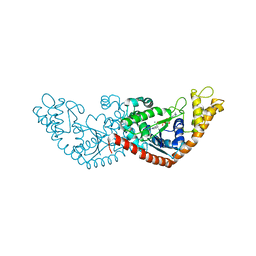 | |
7ENT
 
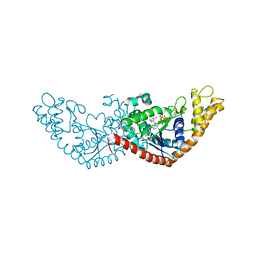 | |
7EV3
 
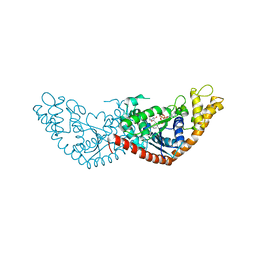 | |
7EV2
 
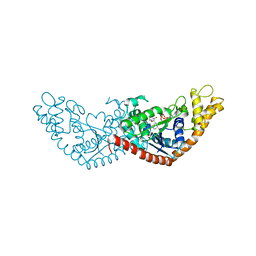 | |
5IY4
 
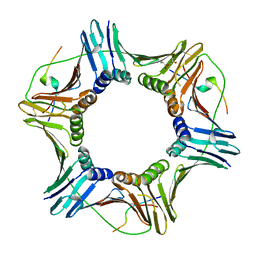 | |
8JCB
 
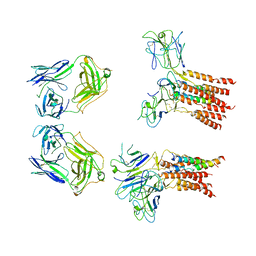 | | Vgamma5 Vdelta1 T cell receptor complex | | Descriptor: | T cell receptor delta variable 1,T cell receptor delta constant, T cell receptor gamma variable 5,T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8YC0
 
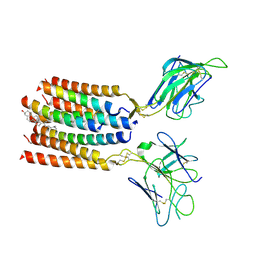 | | T cell receptor V delta2 V gamma9 in GDN | | Descriptor: | CHOLESTEROL, T cell receptor delta variable 2,T cell receptor delta constant, T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2024-02-17 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8JC0
 
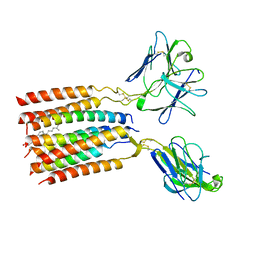 | | V gamma9 V delta2 TCR and CD3 complex in LMNG | | Descriptor: | CHOLESTEROL, T cell receptor delta variable 2,T cell receptor delta constant, T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
1EF0
 
 | |
