9FLW
 
 | | CryoEM structure of the fragment-6 (3571-4078) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9FLS
 
 | | CryoEM structure of the fragment-3 (2061-2397) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9FLY
 
 | | CryoEM structure of the base (4151-5009) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
6LCP
 
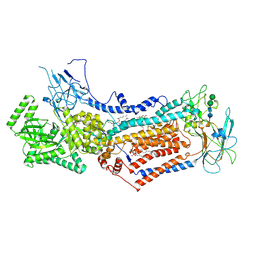 | | Cryo-EM structure of Dnf1 from Chaetomium thermophilum in the E2P state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, Y, Xu, J, Wu, X, Li, L. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structures of a P4-ATPase lipid flippase in lipid bilayers.
Protein Cell, 11, 2020
|
|
6LCR
 
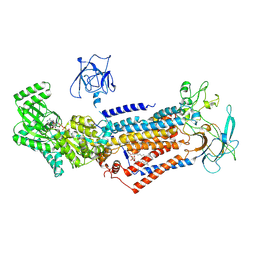 | | Cryo-EM structure of Dnf1 from Chaetomium thermophilum in the E1-ATP state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cdc50, ... | | Authors: | He, Y, Xu, J, Wu, X, Li, L. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a P4-ATPase lipid flippase in lipid bilayers.
Protein Cell, 11, 2020
|
|
8IGN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with RAY1216 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S})-4-(cyclopentylamino)-3,4-bis(oxidanylidene)-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8IGO
 
 | | Crystal structure of apo SARS-CoV-2 main protease | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8GS8
 
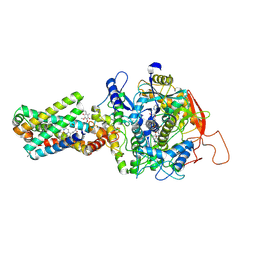 | | cryo-EM structure of the human respiratory complex II | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Du, Z, Zhou, X, Lai, Y, Xu, J, Zhang, Y, Zhou, S, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-09-05 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure of the human respiratory complex II.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9AVA
 
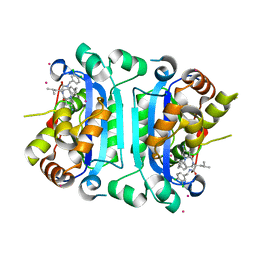 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | (2R)-2-[(5R,6S,8R,9aS)-8-amino-1-oxo-5-(2-phenylethyl)hexahydro-1H-pyrrolo[1,2-a][1,4]diazepin-2(3H)-yl]-N-[(3,4-dichlorophenyl)methyl]-4-methylpentanamide, POTASSIUM ION, Three-prime repair exonuclease 1, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Xu, J, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-01 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
6ZS5
 
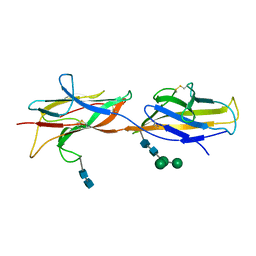 | | 3.5 A cryo-EM structure of human uromodulin filament core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
6ZYA
 
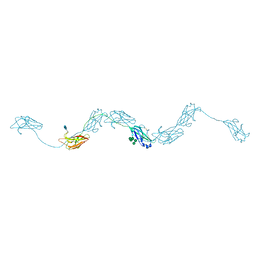 | | Extended human uromodulin filament core at 3.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
8JT3
 
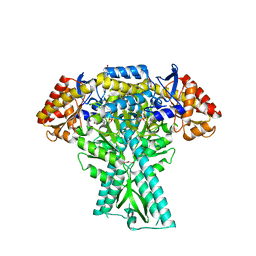 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with amino donor L-Arg | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, ACETATE ION, CrmG, ... | | Authors: | Su, K, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2023-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Co-crystal structure provides insights on transaminase CrmG recognition amino donor L-Arg.
Biochem.Biophys.Res.Commun., 675, 2023
|
|
8HQL
 
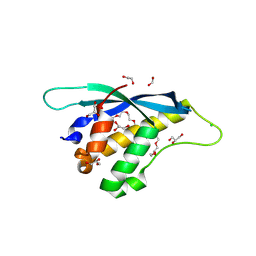 | | Crystal structure of mouse SNX25 PX domain | | Descriptor: | ACETIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yu, Z, Xu, J, Liu, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Redox-modulated SNX25 as a novel regulator of GPCR-G protein signaling from endosomes.
Redox Biol, 75, 2024
|
|
6H67
 
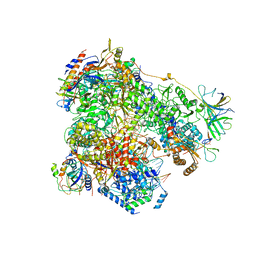 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | Deposit date: | 2018-07-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6H68
 
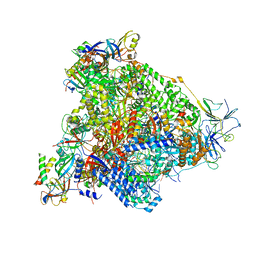 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) with fully-ordered A49 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | Deposit date: | 2018-07-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8HCU
 
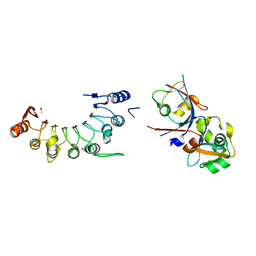 | | Crystal structure of BCOR/PCGF1/KDM2B complex | | Descriptor: | ACETIC ACID, BCL-6 corepressor, Polycomb group RING finger protein 1, ... | | Authors: | Shen, F, Chen, R, Xu, J, Liu, J. | | Deposit date: | 2022-11-03 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Calcium modulates the tethering of BCOR-PRC1.1 enzymatic core to KDM2B via liquid-liquid phase separation.
Commun Biol, 7, 2024
|
|
4PZG
 
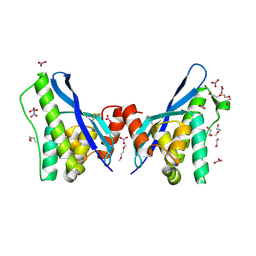 | | Crystal structure of human sorting nexin 10 (SNX10) | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, NITRATE ION, Sorting nexin-10 | | Authors: | Xu, T, Xu, J, Wang, Q, Liu, J. | | Deposit date: | 2014-03-30 | | Release date: | 2014-09-24 | | Last modified: | 2014-12-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human SNX10 reveals insights into its role in human autosomal recessive osteopetrosis.
Proteins, 82, 2014
|
|
5UF7
 
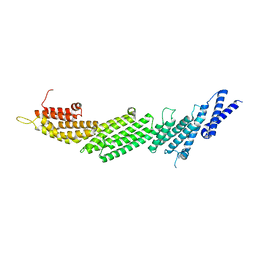 | | CRYSTAL STRUCTURE OF MUNC13-1 MUN DOMAIN | | Descriptor: | Protein unc-13 homolog A | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2017-01-03 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
7WF9
 
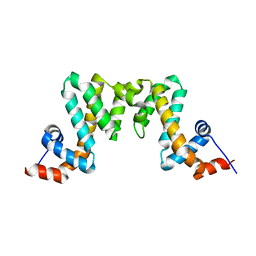 | |
7WF8
 
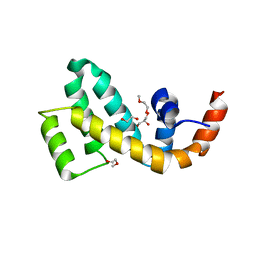 | | Crystal structure of mouse SNX25 RGS domain in space group P212121 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2021-12-26 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Studies Reveal Unique Non-canonical Regulators of G Protein Signaling Homology (RH) Domains in Sorting Nexins.
J.Mol.Biol., 434, 2022
|
|
8GXV
 
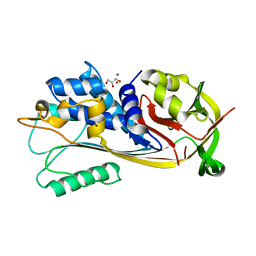 | |
7WPT
 
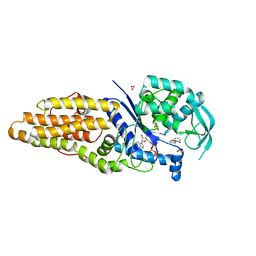 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment M2-80 and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPK
 
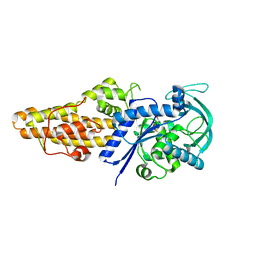 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with L-Met | | Descriptor: | METHIONINE, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPJ
 
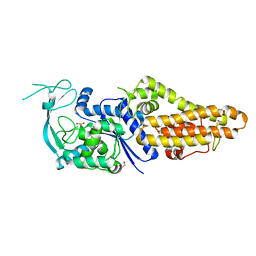 | | Methionyl-tRNA synthetase from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
