8W13
 
 | | Crystal structure of MYST acetyltransferase domain in complex with N-(1-(5-bromo-2-methoxyphenyl)-1H-1,2,3-triazol-4-yl)-2-methoxybenzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone acetyltransferase KAT8, ... | | Authors: | Chen, C, Dou, Y, Wang, M, Xu, C, Buesking, A. | | Deposit date: | 2024-02-15 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identification of triazolyl KAT6 inhibitors via a templated fragment approach.
Bioorg.Med.Chem.Lett., 113, 2024
|
|
3N2C
 
 | | Crystal structure of prolidase eah89906 complexed with n-methylphosphonate-l-proline | | Descriptor: | 1-[(R)-hydroxy(methyl)phosphoryl]-L-proline, PROLIDASE, ZINC ION | | Authors: | Patskovsky, Y, Xu, C, Sauder, J.M, Burley, S.K, Raushel, F.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
8WQD
 
 | |
4JXM
 
 | | Crystal structure of RRP9 WD40 repeats | | Descriptor: | U3 small nucleolar RNA-interacting protein 2, UNKNOWN ATOM OR ION | | Authors: | Wu, X, Tempel, W, Xu, C, El Bakkouri, M, He, H, Seitova, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of RRP9 WD40 repeats
To be Published
|
|
7X5M
 
 | | Tir-dsDNA complex, the initial binding state | | Descriptor: | 2',3'- cyclic AMP, DNA (5'-D(P*AP*TP*TP*AP*A)-3'), DNA (5'-D(P*AP*TP*TP*TP*A)-3'), ... | | Authors: | Tan, Y, Xu, C, Yu, D, Song, W, Wu, B, Schulze-Lefert, P, Chai, J. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | TIR domains of plant immune receptors are 2',3'-cAMP/cGMP synthetases mediating cell death.
Cell, 185, 2022
|
|
3J8X
 
 | | High-resolution structure of no-nucleotide kinesin on microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Shang, Z, Zhou, K, Xu, C, Csencsits, R, Cochran, J.C, Sindelar, C.V. | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation.
Elife, 3, 2014
|
|
2LTP
 
 | | Solution structure of the SANT2 domain of the human nuclear receptor corepressor 2 (NCoR2), Northeast Structural Genomics Consortium (NESG) target ID HR4636E | | Descriptor: | Nuclear receptor corepressor 2 | | Authors: | Montecchio, M, Lemak, A, Yee, A, Xu, C, Garcia, M, Houliston, S, Bellanda, M, Min, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SANT2 domain of the human nuclear receptor corepressor 2 (NCoR2).
To be Published
|
|
4RCJ
 
 | | Crystal structure of YTHDF1 YTH domain in complex with 5mer m6A RNA | | Descriptor: | RNA (5'-R(*GP*GP*(6MZ)P*CP*U)-3'), UNKNOWN ATOM OR ION, YTH domain-containing family protein 1 | | Authors: | Tempel, W, Xu, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
5YBJ
 
 | | Structure of apo KANK1 ankyrin domain | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
6J8F
 
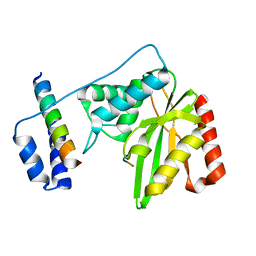 | | Crystal structure of SVBP-VASH1 with peptide mimic the C-terminal of alpha-tubulin | | Descriptor: | 8-mer peptide, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-18 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.283 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
3LX7
 
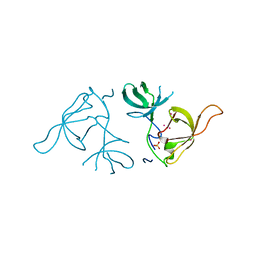 | | Crystal structure of a Novel Tudor domain-containing protein SGF29 | | Descriptor: | SAGA-associated factor 29 homolog, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Bian, C.B, Xu, C, Tempel, W, Lam, R, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J. | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
5YBU
 
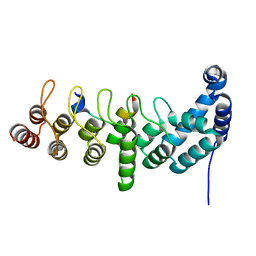 | | Structure of the KANK1 ankyrin domain in complex with KIF21A peptide | | Descriptor: | KN motif and ankyrin repeat domain-containing protein 1, Kinesin-like protein KIF21A | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
3IUF
 
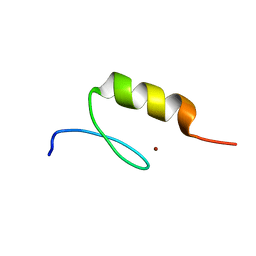 | | Crystal structure of the C2H2-type zinc finger domain of human ubi-d4 | | Descriptor: | ZINC ION, Zinc finger protein ubi-d4 | | Authors: | Tempel, W, Xu, C, Bian, C, Adams-Cioaba, M, Eryilmaz, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Cys2His2-type zinc finger domain of human DPF2.
Biochem.Biophys.Res.Commun., 413, 2011
|
|
3ME9
 
 | | Crystal structure of SGF29 in complex with H3K4me3 peptide | | Descriptor: | GLYCEROL, Histone H3, SAGA-associated factor 29 homolog, ... | | Authors: | Bian, C, Tempel, W, Xu, C, Guo, Y, Dong, A, Crombet, L, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
8WQF
 
 | | cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 2) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQE
 
 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 1) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQG
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQC
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CDK5R1 | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQA
 
 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQB
 
 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQH
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
6CNQ
 
 | | MBD2 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Xu, C, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-08 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA.
J. Biol. Chem., 293, 2018
|
|
6C1A
 
 | | MBD2 in complex with methylated DNA | | Descriptor: | Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION, complement to dna strand 1, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-04 | | Release date: | 2018-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA.
J. Biol. Chem., 293, 2018
|
|
3MET
 
 | | Crystal structure of SGF29 in complex with H3K4me2 | | Descriptor: | GLYCEROL, Histone H3, SAGA-associated factor 29 homolog, ... | | Authors: | Bian, C.B, Xu, C, Lam, R, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
3K27
 
 | | Complex structure of EED and trimethylated H3K9 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Bian, C.B, Xu, C, Qiu, W, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
