1SSW
 
 | | Crystal structure of phage T4 lysozyme mutant Y24A/Y25A/T26A/I27A/C54T/C97A | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme | | Authors: | He, M.M, Baase, W.A, Xiao, H, Heinz, D.W, Matthews, B.W. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation
Protein Sci., 13, 2004
|
|
4F23
 
 | | Influenza A virus hemagglutinin H16 HA0 structure with an alpha-helix conformation in the cleavage site: a potential drug target | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Lu, X, Shi, Y, Gao, F, Xiao, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-07 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into Avian Influenza Virus Pathogenicity: the Hemagglutinin Precursor HA0 of Subtype H16 Has an Alpha-Helix Structure in Its Cleavage Site with Inefficient HA1/HA2 Cleavage.
J.Virol., 86, 2012
|
|
4FIU
 
 | | The structure of hemagglutinin of H16 subtype influenza virus with V327G mutation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Lu, X, Shi, Y, Gao, F, Xiao, H, Wang, M, Qi, J, Gao, G.F. | | Deposit date: | 2012-06-11 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Insights into Avian Influenza Virus Pathogenicity: the Hemagglutinin Precursor HA0 of Subtype H16 Has an Alpha-Helix Structure in Its Cleavage Site with Inefficient HA1/HA2 Cleavage.
J.Virol., 86, 2012
|
|
1T8G
 
 | | Crystal structure of phage T4 lysozyme mutant L32A/L33A/T34A/C54T/C97A/E108V | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | Authors: | He, M.M, Wood, Z.A, Baase, W.A, Xiao, H, Matthews, B.W. | | Deposit date: | 2004-05-12 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation.
Protein Sci., 13, 2004
|
|
1T8F
 
 | | Crystal structure of phage T4 lysozyme mutant R14A/K16A/I17A/K19A/T21A/E22A/C54T/C97A | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | He, M.M, Wood, Z.A, Baase, W.A, Xiao, H, Matthews, B.W. | | Deposit date: | 2004-05-12 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation.
Protein Sci., 13, 2004
|
|
1SSY
 
 | | Crystal structure of phage T4 lysozyme mutant G28A/I29A/G30A/C54T/C97A | | Descriptor: | Lysozyme | | Authors: | He, M.M, Baase, W.A, Xiao, H, Heinz, D.W, Matthews, B.W. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation
Protein Sci., 13, 2004
|
|
5HGU
 
 | |
8HNI
 
 | | hnRNP A2/B1 RRMs in complex with telomeric DNA | | Descriptor: | DNA (5'-D(P*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*T)-3'), Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Liu, Y, Abula, A, Xiao, H, Guo, H, Li, T, Zheng, L, Chen, B, Nguyen, H, Ji, X. | | Deposit date: | 2022-12-07 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Structural Insight Into hnRNP A2/B1 Homodimerization and DNA Recognition.
J.Mol.Biol., 435, 2023
|
|
7UX8
 
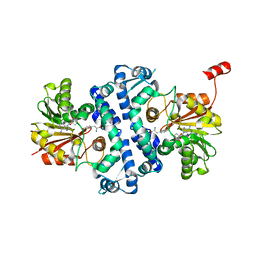 | | Crystal structure of MfnG, an L- and D-tyrosine O-methyltransferase from the marformycin biosynthesis pathway of Streptomyces drozdowiczii, with SAH and L-Tyrosine bound at 1.4 A resolution (P212121 - form II) | | Descriptor: | MfnG, S-ADENOSYL-L-HOMOCYSTEINE, TYROSINE, ... | | Authors: | Miller, M.D, Wu, K.-L, Xu, W, Xiao, H, Philips Jr, G.N. | | Deposit date: | 2022-05-05 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expanding the eukaryotic genetic code with a biosynthesized 21st amino acid.
Protein Sci., 31, 2022
|
|
7UX7
 
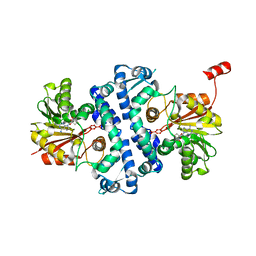 | | Crystal structure of MfnG, an L- and D-tyrosine O-methyltransferase from the marformycin biosynthesis pathway of Streptomyces drozdowiczii, with SAH bound at 1.2 A resolution (P212121 - form II) | | Descriptor: | MfnG, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN LIGAND | | Authors: | Miller, M.D, Wu, K.-L, Xu, W, Xiao, H, Philips Jr, G.N. | | Deposit date: | 2022-05-05 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Expanding the eukaryotic genetic code with a biosynthesized 21st amino acid.
Protein Sci., 31, 2022
|
|
7UX6
 
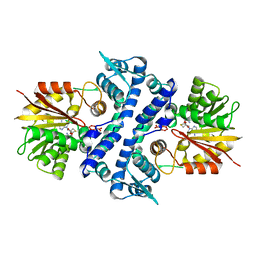 | | Crystal structure of MfnG, an L- and D-tyrosine O-methyltransferase from the marformycin biosynthesis pathway of Streptomyces drozdowiczii, with SAH bound at 1.35 A resolution (P212121 - form I) | | Descriptor: | MfnG, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN LIGAND | | Authors: | Miller, M.D, Wu, K.-L, Xu, W, Xiao, H, Philips Jr, G.N. | | Deposit date: | 2022-05-05 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Expanding the eukaryotic genetic code with a biosynthesized 21st amino acid.
Protein Sci., 31, 2022
|
|
2L5A
 
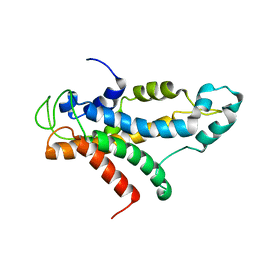 | | Structural basis for recognition of centromere specific histone H3 variant by nonhistone Scm3 | | Descriptor: | Histone H3-like centromeric protein CSE4, Protein SCM3, Histone H4 | | Authors: | Zhou, Z, Feng, H, Zhou, B, Ghirlando, R, Hu, K, Zwolak, A, Jenkins, L, Xiao, H, Tjandra, N, Wu, C, Bai, Y. | | Deposit date: | 2010-10-28 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3.
Nature, 472, 2011
|
|
4GQ0
 
 | | Crystal structure of AKR1B10 complexed with NADP+ and Caffeic acid phenethyl ester | | Descriptor: | 2-phenylethyl (2E)-3-(3,4-dihydroxyphenyl)prop-2-enoate, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liping, Z, Xuehua, Z, Shangke, C, Jing, Z, Xiaopeng, H. | | Deposit date: | 2012-08-22 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of AKR1B10 complexed with NADP+ and Caffeic acid phenethyl ester
To be Published
|
|
