3WP1
 
 | | Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl | | Descriptor: | Disks large homolog 4, Lethal(2) giant larvae protein homolog 2, SULFATE ION | | Authors: | Zhu, J, Shang, Y, Wan, Q, Xia, Y, Chen, J, Du, Q, Zhang, M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl
Cell Res., 24, 2014
|
|
3WP0
 
 | | Crystal structure of Dlg GK in complex with a phosphor-Lgl2 peptide | | Descriptor: | Disks large homolog 4, GLYCEROL, Lethal(2) giant larvae protein homolog 2 | | Authors: | Zhu, J, Shang, Y, Wan, Q, Xia, Y, Chen, J, Du, Q, Zhang, M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-19 | | Last modified: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl
Cell Res., 24, 2014
|
|
2HEM
 
 | | NMR structure and Mg2+ binding of an RNA segment that underlies the L7/L12 stalk in the E.coli 50S ribosomal subunit. | | Descriptor: | 5'-R(P*GP*GP*GP*AP*AP*GP*GP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*CP*GP*GP*CP*CP*C)-3' | | Authors: | Zhao, Q, Nagaswamy, U, Lee, H, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2006-06-21 | | Release date: | 2006-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and Mg2+ binding of an RNA segment that underlies the L7/L12 stalk in the E.coli 50S ribosomal subunit
Nucleic Acids Res., 33, 2005
|
|
1IAY
 
 | | CRYSTAL STRUCTURE OF ACC SYNTHASE COMPLEXED WITH COFACTOR PLP AND INHIBITOR AVG | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE 2, 2-AMINO-4-(2-AMINO-ETHOXY)-BUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Huai, Q, Xia, Y, Chen, Y, Callahan, B, Li, N, Ke, H. | | Deposit date: | 2001-03-24 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of 1-aminocyclopropane-1-carboxylate (ACC) synthase in complex with aminoethoxyvinylglycine and pyridoxal-5'-phosphate provide new insight into catalytic mechanisms
J.Biol.Chem., 276, 2001
|
|
4N1Z
 
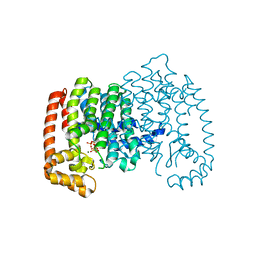 | | Crystal Structure of Human Farnesyl Diphosphate Synthase in Complex with BPH-1222 | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Liu, Y.L, Xia, Y, Zhang, Y, Verma, I, Oldfield, E. | | Deposit date: | 2013-10-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A combination therapy for KRAS-driven lung adenocarcinomas using lipophilic bisphosphonates and rapamycin.
Sci Transl Med, 6, 2014
|
|
4LQT
 
 | | 1.10A resolution crystal structure of a superfolder green fluorescent protein (W57A) mutant | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein | | Authors: | Lovell, S, Xia, Y, Vo, B, Battaile, K.P, Egan, C, Karanicolas, J. | | Deposit date: | 2013-07-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The designability of protein switches by chemical rescue of structure: mechanisms of inactivation and reactivation.
J.Am.Chem.Soc., 135, 2013
|
|
4LQU
 
 | | 1.60A resolution crystal structure of a superfolder green fluorescent protein (W57G) mutant | | Descriptor: | Green fluorescent protein | | Authors: | Lovell, S, Xia, Y, Vo, B, Battaile, K.P, Egan, C, Karanicolas, J. | | Deposit date: | 2013-07-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The designability of protein switches by chemical rescue of structure: mechanisms of inactivation and reactivation.
J.Am.Chem.Soc., 135, 2013
|
|
4I0U
 
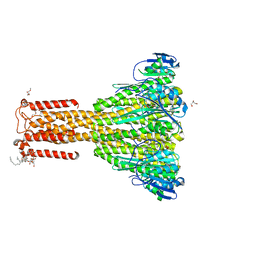 | | Improved structure of Thermotoga maritima CorA at 2.7 A resolution | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nordin, N, Guskov, A, Phua, T, Sahaf, N, Xia, Y, Lu, S.Y, Eshaghi, H, Eshaghi, S. | | Deposit date: | 2012-11-19 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploring the structure and function of Thermotoga maritima CorA reveals the mechanism of gating and ion selectivity in Co2+/Mg2+ transport.
Biochem.J., 451, 2013
|
|
8GVC
 
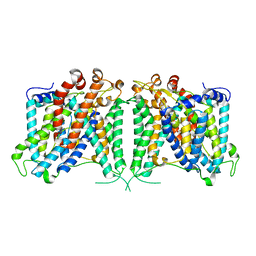 | | The cryo-EM structure of hAE2 with bicarbonate | | Descriptor: | Anion exchange protein 2, BICARBONATE ION | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVF
 
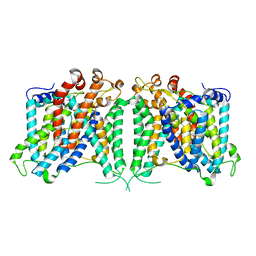 | | The outward-facing structure of hAE2 in basic pH | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVH
 
 | | Human AE2 in acidic KNO3 | | Descriptor: | Anion exchange protein 2, CHOLESTEROL HEMISUCCINATE | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GV9
 
 | | The cryo-EM structure of hAE2 with chloride ion | | Descriptor: | Anion exchange protein 2, CHLORIDE ION | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GV8
 
 | | The cryo-EM structure of hAE2 with DIDS | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVA
 
 | | The intermediate structure of hAE2 in basic pH | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVE
 
 | | The asymmetry structure of hAE2 | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
6JMU
 
 | | Crystal structure of GIT1/Paxillin complex | | Descriptor: | ARF GTPase-activating protein GIT1, Paxillin | | Authors: | Zhu, J, Lin, L, Xia, Y, Zhang, R, Zhang, M. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling.
Mol.Cell, 79, 2020
|
|
6JMT
 
 | | Crystal structure of GIT/PIX complex | | Descriptor: | ARF GTPase-activating protein GIT2, ZINC ION, beta PIX | | Authors: | Zhu, J, Lin, L, Xia, Y, Zhang, R, Zhang, M. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling.
Mol.Cell, 79, 2020
|
|
2MZ0
 
 | |
8JFQ
 
 | | Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-back Loop | | Descriptor: | 26mer-DNA | | Authors: | Liu, Y, Li, J, Zhang, Y, Wang, Y, Chen, J, Bian, Y, Xia, Y, Yang, M.H, Zheng, K, Wang, K.B, Kong, L.Y. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-Back Loop.
J.Am.Chem.Soc., 145, 2023
|
|
2N9B
 
 | |
7CZB
 
 | | The cryo-EM structure of the ERAD retrotranslocation channel formed by human Derlin-1 | | Descriptor: | Derlin-1 | | Authors: | Rao, B, Li, S, Yao, D, Wang, Q, Xia, Y, Jia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2020-09-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The cryo-EM structure of an ERAD protein channel formed by tetrameric human Derlin-1.
Sci Adv, 7, 2021
|
|
7F3X
 
 | | Lysophospholipid acyltransferase LPCAT3 in complex with lysophosphatidylcholine | | Descriptor: | LPCAT3, [2-((1-OXODODECANOXY-(2-HYDROXY-3-PROPANYL))-PHOSPHONATE-OXY)-ETHYL]-TRIMETHYLAMMONIUM | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
7F40
 
 | | Lysophospholipid acyltransferase LPCAT3 in a complex with Arachidonoyl-CoA | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, LPCAT3, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenethioate | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
7EWT
 
 | | The crystal structure of Lysophospholipid acyltransferase LPCAT3 (MOBAT5) in its monomeric and apo form | | Descriptor: | Lysophospholipid acyltransferase 5 | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
8CYI
 
 | | Cryo-EM structures and computational analysis for enhanced potency in MTA-synergic inhibition of human protein arginine methyltransferase 5 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, N-[(2-aminoquinolin-7-yl)methyl]-9-(2-hydroxyethyl)-2,3,4,9-tetrahydro-1H-carbazole-6-carboxamide, ... | | Authors: | Yadav, G.P, Wei, Z, Xiaozhi, Y, Chenglong, L, Jiang, Q. | | Deposit date: | 2022-05-23 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structure-based selection of computed ligand poses enables design of MTA-synergic PRMT5 inhibitors of better potency.
Commun Biol, 5, 2022
|
|
