1G70
 
 | | COMPLEX OF HIV-1 RRE-IIB RNA WITH RSG-1.2 PEPTIDE | | Descriptor: | HIV-1 RRE-IIB 32 NUCLEOTIDE RNA, RSG-1.2 PEPTIDE | | Authors: | Gosser, Y, Hermann, T, Majumdar, A, Hu, W, Frederick, R, Jiang, F, Xu, W, Patel, D.J. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Peptide-triggered conformational switch in HIV-1 RRE RNA complexes.
Nat.Struct.Biol., 8, 2001
|
|
2Z6H
 
 | | Crystal Structure of Beta-Catenin Armadillo Repeat Region and Its C-Terminal domain | | Descriptor: | Catenin beta-1 | | Authors: | Xing, Y, Takemaru, K, Liu, J, Zheng, J, Moon, R, Xu, W. | | Deposit date: | 2007-08-01 | | Release date: | 2008-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Full-Length beta-Catenin
Structure, 16, 2008
|
|
1LUJ
 
 | | Crystal Structure of the Beta-catenin/ICAT Complex | | Descriptor: | Beta-catenin-interacting protein 1, Catenin beta-1 | | Authors: | Graham, T.A, Clements, W.K, Kimelman, D, Xu, W. | | Deposit date: | 2002-05-22 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the beta-catenin/ICAT complex reveals the inhibitory mechanism of ICAT.
Mol.Cell, 10, 2002
|
|
8K47
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K45
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb4 nanobody, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
6VJV
 
 | | Crystal structure of the Prochlorococcus phage (myovirus P-SSM2) ferredoxin at 1.6 Angstroms | | Descriptor: | ACETATE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | Authors: | Olmos Jr, J.L, Campbell, I.J, Miller, M.D, Xu, W, Kahanda, D, Atkinson, J.T, Sparks, N, Bennett, G.N, Silberg, J.J, Phillips Jr, G.N. | | Deposit date: | 2020-01-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Prochlorococcusphage ferredoxin: structural characterization and electron transfer to cyanobacterial sulfite reductases.
J.Biol.Chem., 295, 2020
|
|
1QZ7
 
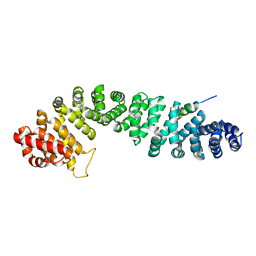 | | Beta-catenin binding domain of Axin in complex with beta-catenin | | Descriptor: | Axin, Beta-catenin | | Authors: | Xing, Y, Clements, W.K, Kimelman, D, Xu, W. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a beta-catenin/Axin complex suggests a mechanism for the {beta}-catenin destruction complex
GENES DEV., 17, 2003
|
|
1XDO
 
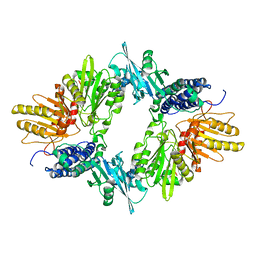 | |
1Z87
 
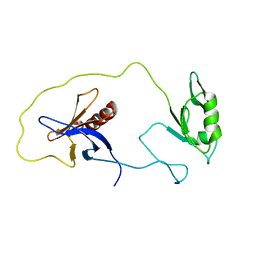 | | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | | Descriptor: | Alpha-1-syntrophin | | Authors: | Yan, J, Xu, W, Wen, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | Deposit date: | 2005-03-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
1Z86
 
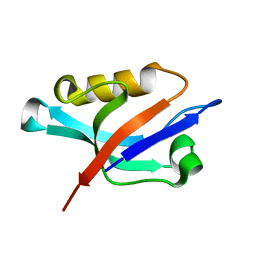 | | Solution structure of the PDZ domain of alpha-syntrophin | | Descriptor: | Alpha-1-syntrophin | | Authors: | Yan, J, Xu, W, Wen, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | Deposit date: | 2005-03-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
1XDP
 
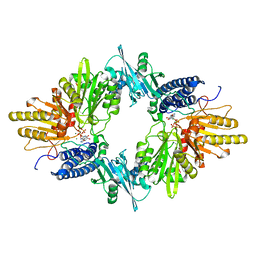 | | Crystal Structure of the E.coli Polyphosphate Kinase in complex with AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyphosphate kinase | | Authors: | Zhu, Y, Huang, W, Lee, S.S, Xu, W. | | Deposit date: | 2004-09-07 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a polyphosphate kinase and its implications for polyphosphate synthesis
Embo Rep., 6, 2005
|
|
1NQZ
 
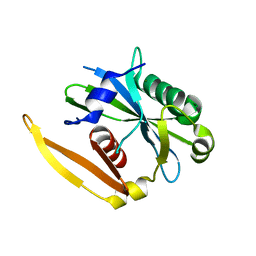 | | The structure of a CoA pyrophosphatase from D. Radiodurans complexed with a magnesium ion | | Descriptor: | CoA pyrophosphatase (MutT/nudix family protein), MAGNESIUM ION | | Authors: | Kang, L.W, Gabelli, S.B, Bianchet, M.A, Xu, W.L, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family.
J.Bacteriol., 185, 2003
|
|
2ADZ
 
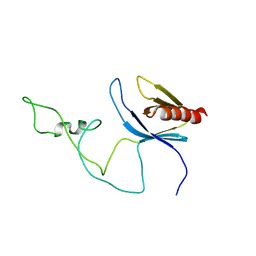 | | solution structure of the joined PH domain of alpha1-syntrophin | | Descriptor: | Alpha-1-syntrophin | | Authors: | Yan, J, Wen, W, Xu, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
1NQY
 
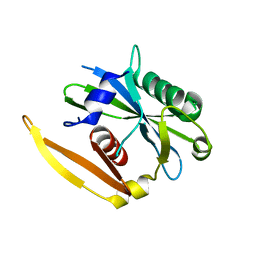 | | The structure of a CoA pyrophosphatase from D. Radiodurans | | Descriptor: | CoA pyrophosphatase (MutT/nudix family protein) | | Authors: | Kang, L.W, Gabelli, S.B, Bianchet, M.A, Xu, W.L, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family.
J.Bacteriol., 185, 2003
|
|
1NEM
 
 | | Saccharide-RNA recognition in the neomycin B / RNA aptamer complex | | Descriptor: | 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranose, 2,6-diamino-2,6-dideoxy-beta-L-idopyranose-(1-3)-beta-D-ribofuranose, 2-DEOXY-D-STREPTAMINE, ... | | Authors: | Jiang, L, Majumdar, A, Hu, W, Jaishree, T.J, Xu, W, Patel, D.J. | | Deposit date: | 1999-03-15 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in a complex formed between neomycin B and an RNA aptamer
Structure Fold.Des., 7, 1999
|
|
1JDH
 
 | | CRYSTAL STRUCTURE OF BETA-CATENIN AND HTCF-4 | | Descriptor: | BETA-CATENIN, hTcf-4 | | Authors: | Graham, T.A, Ferkey, D.M, Mao, F, Kimelman, D, Xu, W. | | Deposit date: | 2001-06-13 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tcf4 can specifically recognize beta-catenin using alternative conformations.
Nat.Struct.Biol., 8, 2001
|
|
6UBL
 
 | | Structure of DynF from the Dynemicin Biosynthesis Pathway of Micromonospora chersina | | Descriptor: | DynF, PALMITIC ACID | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Bhardwaj, M, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6P9V
 
 | | Crystal Structure of hMAT Mutant K289L | | Descriptor: | ADENOSINE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Miller, M.D, Xu, W, Huber, T.D, Clinger, J.A, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Methionine Adenosyltransferase Engineering to Enable Bioorthogonal Platforms for AdoMet-Utilizing Enzymes.
Acs Chem.Biol., 15, 2020
|
|
1SJY
 
 | | Crystal Structure of NUDIX HYDROLASE DR1025 FROM DEINOCOCCUS RADIODURANS | | Descriptor: | MutT/nudix family protein | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
6VQP
 
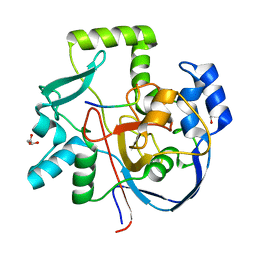 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CalU17, CalU17 His-Tagged protein, GLYCEROL, ... | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of DynF from the Dynemicin Biosynthesis Pathway of Micromonospora chersina
To Be Published
|
|
1SOI
 
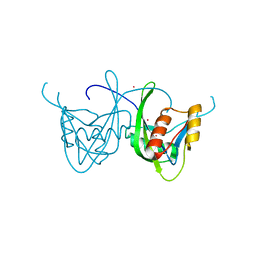 | | CRYSTAL STRUCTURE OF NUDIX HYDROLASE DR1025 IN COMPLEX WITH SM+3 | | Descriptor: | MutT/nudix family protein, SAMARIUM (III) ION | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-15 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
7VUX
 
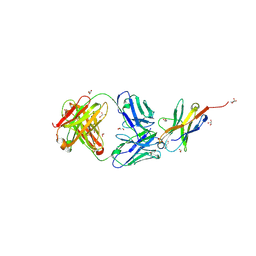 | | Complex structure of PD1 and 609A-Fab | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Huang, H, Zhu, Z, Zhao, J, Jiang, L, Yang, H, Deng, L, Meng, X, Ding, J, Yang, S, Zhao, L, Xu, W, Wang, X. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A strategy for the efficient construction of anti-PD1-based bispecific antibodies with desired IgG-like properties.
Mabs, 14, 2022
|
|
2AZR
 
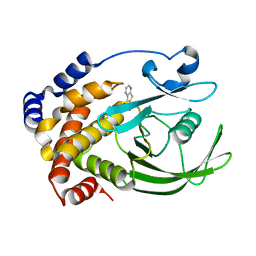 | | Crystal structure of PTP1B with Bicyclic Thiophene inhibitor | | Descriptor: | 3-(CARBOXYMETHOXY)THIENO[2,3-B]PYRIDINE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase, non-receptor type 1 | | Authors: | Moretto, A.F, Kirincich, S.J, Xu, W.X, Smith, M.J, Wan, Z.K, Wilson, D.P, Follows, B.C, Binnun, E, Joseph-McCarthy, D, Foreman, K, Erbe, D.V, Zhang, Y.L, Tam, S.K, Tam, S.Y, Lee, J. | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bicyclic and tricyclic thiophenes as protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem., 14, 2006
|
|
2B07
 
 | | Crystal structure of PTP1B with Tricyclic Thiophene inhibitor. | | Descriptor: | 6-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}-3-(CARBOXYMETHOXY)THIENO[3,2-B][1]BENZOTHIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase, non-receptor type 1 | | Authors: | Moretto, A.F, Kirincich, S.J, Xu, W.X, Smith, M.J, Wan, Z.K, Wilson, D.P, Follows, B.C, Binnun, E, Joseph-McCarthy, D, Foreman, K, Erbe, D.V, Zhang, Y.L, Tam, S.K, Tam, S.Y, Lee, J. | | Deposit date: | 2005-09-13 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bicyclic and tricyclic thiophenes as protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem., 14, 2006
|
|
