1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
2FPL
 
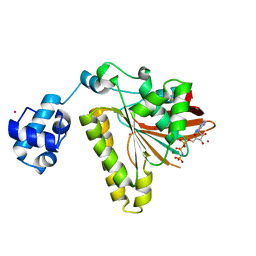 | | RadA recombinase in complex with AMP-PNP and low concentration of K+ | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Methanococcus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
2FPM
 
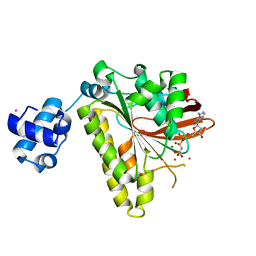 | | RadA recombinase in complex with AMP-PNP and high concentration of K+ | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Methanoccocus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
2FPK
 
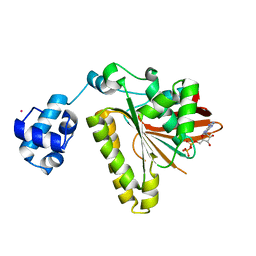 | | RadA recombinase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair and recombination protein radA, MAGNESIUM ION, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Methanococcus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
3KOV
 
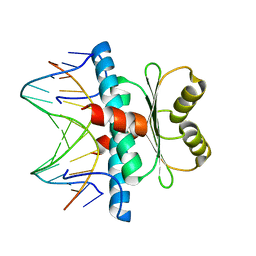 | | Structure of MEF2A bound to DNA reveals a completely folded MADS-box/MEF2 domain that recognizes DNA and recruits transcription co-factors | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), Myocyte-specific enhancer factor 2A | | Authors: | Wu, Y, Dey, R, Han, A, Jayathilaka, N, Philips, M, Ye, J, Chen, L. | | Deposit date: | 2009-11-14 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the MADS-box/MEF2 Domain of MEF2A Bound to DNA and Its Implication for Myocardin Recruitment.
J.Mol.Biol., 397, 2010
|
|
4MWV
 
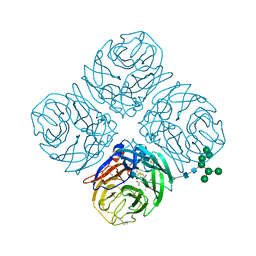 | | Anhui N9-peramivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWL
 
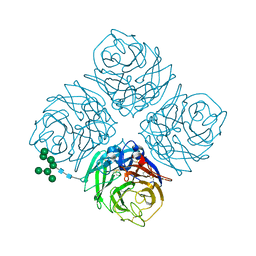 | | Shanghai N9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MX0
 
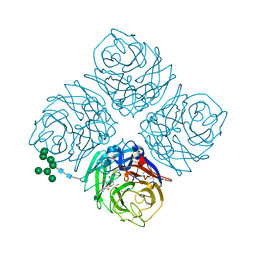 | | Shanghai N9-peramivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWU
 
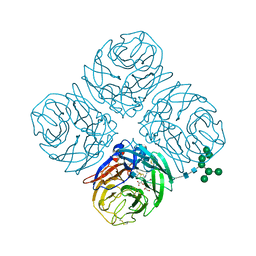 | | Anhui N9-laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWY
 
 | | Shanghai N9-laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
3TZN
 
 | |
4NHG
 
 | | Crystal Structure of 2G12 IgG Dimer | | Descriptor: | 2G12 IgG dimer heavy chain, 2G12 IgG dimer light chain, Hepatitis B virus receptor binding protein | | Authors: | Wu, Y, West Jr, A.P, Kim, H.J, Thornton, M.E, Ward, A.B, Bjorkman, P.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-26 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (8.001 Å) | | Cite: | Structural basis for enhanced HIV-1 neutralization by a dimeric immunoglobulin G form of the glycan-recognizing antibody 2G12.
Cell Rep, 5, 2013
|
|
5J97
 
 | |
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | Authors: | Wu, Y, Qi, J, Gao, F. | | Deposit date: | 2020-04-26 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
8HKH
 
 | |
4RZV
 
 | | Crystal structure of the BRAF (R509H) kinase domain monomer bound to Vemurafenib | | Descriptor: | N-(3-{[5-(4-chlorophenyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Wu, Y, Gavathiotis, E. | | Deposit date: | 2014-12-24 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | An integrated model of RAF inhibitor action predicts
inhibitor activity against oncogenic BRAF signaling
Cancer Cell, 30, 2016
|
|
4RZW
 
 | | Crystal structure of BRAF (R509H) kinase domain bound to AZ628 | | Descriptor: | 3-(2-cyanopropan-2-yl)-N-{4-methyl-3-[(3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]phenyl}benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Wu, Y, Gavathiotis, E. | | Deposit date: | 2014-12-24 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.493 Å) | | Cite: | An integrated model of RAF inhibitor action predicts
inhibitor activity against oncogenic BRAF signaling
Cancer Cell, 30, 2016
|
|
6F5E
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_10_47 in complex JNK1a1 and JIP1 peptide | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, DD_D12_10_47, Mitogen-activated protein kinase 8 | | Authors: | Wu, Y, Mittl, P.R, Honegger, A, Batyuk, A, Plueckthun, A. | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_10_47 in complex JNK1a1 and JIP1 peptide
To be published
|
|
6G8U
 
 | |
6G8V
 
 | |
5BOF
 
 | | Crystal Structure of Staphylococcus aureus Enolase | | Descriptor: | Enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Wu, Y.F, Wang, C.L, Wu, M.H, Han, L, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WBE
 
 | |
5NXQ
 
 | |
4WBP
 
 | |
5LW1
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_232_11_D12 in complex JNK1a1 and JIP1 peptide | | Descriptor: | ADENOSINE, C-Jun-amino-terminal kinase-interacting protein 1, DD_232_11_D12, ... | | Authors: | Wu, Y, Batyuk, A, Mittl, P.R, Honegger, A, Plueckthun, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for the Selective Inhibition of c-Jun N-Terminal Kinase 1 Determined by Rigid DARPin-DARPin Fusions.
J.Mol.Biol., 430, 2018
|
|
