7DHS
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1R)-1-phenylethyl]benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Wu, T, Xiang, Q, Wang, C, Wu, C, Zhang, C, Zhang, M, Liu, Z, Zhang, Y, Xiao, L, Xu, Y. | | Deposit date: | 2020-11-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Y06014 is a selective BET inhibitor for the treatment of prostate cancer.
Acta Pharmacol.Sin., 42, 2021
|
|
1PKR
 
 | |
6KLA
 
 | | Crystal structure of human c-KIT kinase domain in complex with compound 15a | | Descriptor: | Mast/stem cell growth factor receptor Kit, N-[6-(4-ethylpiperazin-1-yl)-2-methyl-pyrimidin-4-yl]-5-pyridin-4-yl-1,3-thiazol-2-amine | | Authors: | Wu, T.S, Peng, Y.H, Hsueh, C.C, Wu, S.Y. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Identification of a Multitargeted Tyrosine Kinase Inhibitor for the Treatment of Gastrointestinal Stromal Tumors and Acute Myeloid Leukemia.
J.Med.Chem., 62, 2019
|
|
4TSR
 
 | |
3AZR
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZS
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZT
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMC
 
 | | Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMG
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AOF
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Mannotriose substrate | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-09-27 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMD
 
 | | Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
8I7H
 
 | | Meso-Diaminopimelate dehydrogenase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Meso-diaminopimelate D-dehydrogenase, SULFATE ION | | Authors: | Wu, T.F, Song, W. | | Deposit date: | 2023-01-31 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Analysis of the catalytic mechanism of meso-DAPDH and extension of D-aromatic amino acid substrate scope
To Be Published
|
|
6ITT
 
 | | Crystal structure of unactivated c-KIT in complex with compound | | Descriptor: | Mast/stem cell growth factor receptor Kit, N-(5-ethyl-1,2-oxazol-3-yl)-N'-[4-(2-{[6-(4-ethylpyrazin-1(4H)-yl)pyrimidin-4-yl]amino}-1,3-thiazol-5-yl)phenyl]urea | | Authors: | Wu, T.S, Wu, S.Y. | | Deposit date: | 2018-11-26 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Crystal structure of unactivated c-KIT in complex with compound
To Be Published
|
|
6ITV
 
 | | Crystal structure of activated c-KIT in complex with compound | | Descriptor: | Mast/stem cell growth factor receptor Kit, N-(5-ethyl-1,2-oxazol-3-yl)-N'-[4-(2-{[6-(4-ethylpyrazin-1(4H)-yl)pyrimidin-4-yl]amino}-1,3-thiazol-5-yl)phenyl]urea | | Authors: | Wu, T.S, Wu, S.Y. | | Deposit date: | 2018-11-26 | | Release date: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Crystal structure of activated c-KIT in complex with compound
To Be Published
|
|
3MRU
 
 | | Crystal Structure of Aminoacylhistidine Dipeptidase from Vibrio alginolyticus | | Descriptor: | Aminoacyl-histidine dipeptidase, ZINC ION | | Authors: | Chang, C.-Y, Hsieh, Y.-C, Wu, T.-K, Chen, C.-J. | | Deposit date: | 2010-04-29 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure and mutational analysis of aminoacylhistidine dipeptidase from vibrio alginolyticus reveal a new architecture of M20 metallopeptidases
J.Biol.Chem., 285, 2010
|
|
4UMP
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 3-(isoquinolin-7-yl)prop-2-yn-1-ol, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4UMR
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 4-fluoro-N-(1,2,3,4-tetrahydroisoquinolin-7-yl)benzamide, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4UMQ
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 3-{5-[(3-hydroxy-5-methoxyphenyl)amino]-2-(phenylcarbamoyl)phenoxy}propan-1-aminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4UMT
 
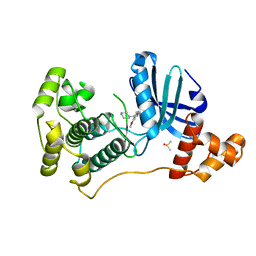 | | Structure of MELK in complex with inhibitors | | Descriptor: | 1-(4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}-2-methoxybenzyl)piperazinediium, DIMETHYL SULFOXIDE, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
4UMU
 
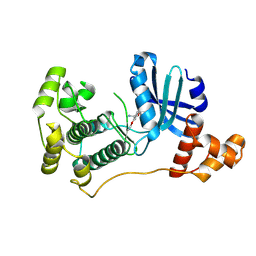 | | Structure of MELK in complex with inhibitors | | Descriptor: | (2-ethoxy-4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}phenyl)methanaminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
3WDU
 
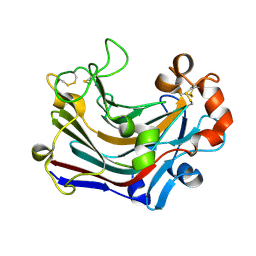 | | The complex structure of PtLic16A with cellobiose | | Descriptor: | Beta-1,3-1,4-glucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WDT
 
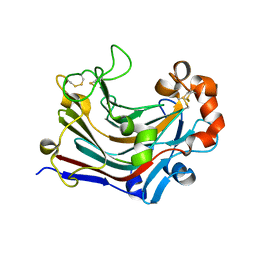 | | The apo-form structure of PtLic16A from Paecilomyces thermophila | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WH9
 
 | | The ligand-free structure of ManBK from Aspergillus niger BK01 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, J.W, Chen, C.C, Huang, C.H, Huang, T.Y, Wu, T.H, Cheng, Y.S, Ko, T.P, Lin, C.Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Analysis and Rational Design to Improve Specific Activity of beta-Mannanase from Aspergillus Niger BK01
To be Published
|
|
3WDY
 
 | | The complex structure of E113A with cellotetraose | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION, beta-D-glucopyranose, ... | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WDW
 
 | | The apo-form structure of E113A from Paecilomyces thermophila | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
