4JBQ
 
 | | Novel Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules | | Descriptor: | Aurora Kinase A, CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE | | Authors: | Wu, J.S, Leou, J.S, Peng, Y.H, Hsueh, C.C, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2013-02-20 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JBP
 
 | | Novel Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules | | Descriptor: | 1-(4-{2-[(6-{4-[2-(4-hydroxypiperidin-1-yl)ethoxy]phenyl}furo[2,3-d]pyrimidin-4-yl)amino]ethyl}phenyl)-3-phenylurea, Aurora Kinase A | | Authors: | Wu, J.S, Leou, J.S, Peng, Y.H, Hsueh, C.C, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2013-02-20 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1JC6
 
 | | SOLUTION STRUCTURE OF BUNGARUS FACIATUS IX, A KUNITZ-TYPE CHYMOTRYPSIN INHIBITOR | | Descriptor: | VENOM BASIC PROTEASE INHIBITORS IX AND VIIIB | | Authors: | Chen, C, Hsu, C.H, Su, N.Y, Chiou, S.H, Wu, S.H. | | Deposit date: | 2001-06-08 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Kunitz-type chymotrypsin inhibitor isolated from the elapid snake Bungarus fasciatus
J.BIOL.CHEM., 276, 2001
|
|
2ATH
 
 | | Crystal structure of the ligand binding domain of human PPAR-gamma im complex with an agonist | | Descriptor: | 2-{5-[3-(7-PROPYL-3-TRIFLUOROMETHYLBENZO[D]ISOXAZOL-6-YLOXY)PROPOXY]INDOL-1-YL}ETHANOIC ACID, Peroxisome proliferator activated receptor gamma | | Authors: | Mahindroo, N, Huang, C.-F, Wu, S.-Y, Hsieh, H.-P. | | Deposit date: | 2005-08-25 | | Release date: | 2006-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Novel indole-based peroxisome proliferator-activated receptor agonists: design, SAR, structural biology, and biological activities
J.Med.Chem., 48, 2005
|
|
5F8W
 
 | | Crystal structure of a Crenomytilus grayanus lectin in complex with galactose | | Descriptor: | GLYCEROL, GalNAc/Gal-specific lectin, alpha-D-galactopyranose, ... | | Authors: | Liao, J.-H, Huang, K.-F, Tu, I.-F, Lee, I.-M, Wu, S.-H. | | Deposit date: | 2015-12-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A Multivalent Marine Lectin from Crenomytilus grayanus Possesses Anti-cancer Activity through Recognizing Globotriose Gb3
J.Am.Chem.Soc., 138, 2016
|
|
5F8S
 
 | | Crystal structure of a Crenomytilus grayanus lectin | | Descriptor: | GLYCEROL, GalNAc/Gal-specific lectin | | Authors: | Liao, J.-H, Huang, K.-F, Tu, I.-F, Lee, I.-M, Wu, S.-H. | | Deposit date: | 2015-12-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | A Multivalent Marine Lectin from Crenomytilus grayanus Possesses Anti-cancer Activity through Recognizing Globotriose Gb3
J.Am.Chem.Soc., 138, 2016
|
|
5F8Y
 
 | | Crystal structure of a Crenomytilus grayanus lectin in complex with galactosamine | | Descriptor: | 2-amino-2-deoxy-alpha-D-galactopyranose, GLYCEROL, GalNAc/Gal-specific lectin | | Authors: | Liao, J.-H, Huang, K.-F, Tu, I.-F, Lee, I.-M, Wu, S.-H. | | Deposit date: | 2015-12-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Multivalent Marine Lectin from Crenomytilus grayanus Possesses Anti-cancer Activity through Recognizing Globotriose Gb3
J.Am.Chem.Soc., 138, 2016
|
|
5F90
 
 | | Crystal structure of a Crenomytilus grayanus lectin in complex with Gb3 allyl | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 2-Propen-1-ol, GalNAc/Gal-specific lectin, ... | | Authors: | Liao, J.-H, Huang, K.-F, Tu, I.-F, Lee, I.-M, Wu, S.-H. | | Deposit date: | 2015-12-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Multivalent Marine Lectin from Crenomytilus grayanus Possesses Anti-cancer Activity through Recognizing Globotriose Gb3
J.Am.Chem.Soc., 138, 2016
|
|
1H8V
 
 | | The X-ray Crystal Structure of the Trichoderma reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Shaw, A, Ropp, T.H, Wu, S, Bott, R, Cameron, A.D, Stahlberg, J, Mitchinson, C, Jones, T.A. | | Deposit date: | 2001-02-16 | | Release date: | 2001-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-Ray Crystal Structure of the Trichoderma Reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution
J.Mol.Biol., 308, 2001
|
|
1XHH
 
 | | Solution Structure of porcine beta-microseminoprotein | | Descriptor: | beta-microseminoprotein | | Authors: | Wang, I, Lou, Y.C, Wu, K.P, Wu, S.H, Chang, W.C, Chen, C. | | Deposit date: | 2004-09-20 | | Release date: | 2005-03-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Novel solution structure of porcine beta-microseminoprotein
J.Mol.Biol., 346, 2005
|
|
2F4B
 
 | | Crystal structure of the ligand binding domain of human PPAR-gamma in complex with an agonist | | Descriptor: | (5-{3-[(6-BENZOYL-1-PROPYL-2-NAPHTHYL)OXY]PROPOXY}-1H-INDOL-1-YL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Lu, I.L, Peng, Y.H, Mahindroo, N, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2005-11-23 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Indol-1-yl Acetic Acids as Peroxisome Proliferator-Activated Receptor Agonists: Design, Synthesis, Structural Biology, and Molecular Docking Studies
J.Med.Chem., 49, 2006
|
|
2G0G
 
 | | Structure-based drug design of a novel family of PPAR partial agonists: virtual screening, x-ray crystallography and in vitro/in vivo biological activities | | Descriptor: | 3-FLUORO-N-[1-(4-FLUOROPHENYL)-3-(2-THIENYL)-1H-PYRAZOL-5-YL]BENZENESULFONAMIDE, Peroxisome proliferator-activated receptor gamma | | Authors: | Lu, I.L, Peng, Y.H, Huang, C.F, Lin, Y.T, Hsu, J.T.A, Wu, S.Y. | | Deposit date: | 2006-02-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure-Based Drug Design of a Novel Family of PPARgamma Partial Agonists: Virtual Screening, X-ray Crystallography, and in Vitro/in Vivo Biological Activities
J.Med.Chem., 49, 2006
|
|
6XNX
 
 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Strand Transfer Complex (Dynamic-Form) | | Descriptor: | 12RSS integration strand DNA (55-MER), 12RSS signal top strand DNA (34-MER), 23RSS integration strand DNA (66-MER), ... | | Authors: | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
6XNZ
 
 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Target Capture Complex | | Descriptor: | 12RSS integration strand (34-mer), 12RSS non-integration strand (34-mer), 23RSS integration strand (45-mer), ... | | Authors: | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
6XNY
 
 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Strand Transfer Complex (Paired-Form) | | Descriptor: | 12RSS integration strand (55-mer), 12RSS signal DNA top strand (34-mer), 23RSS integration strand (66-mer), ... | | Authors: | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
1KU9
 
 | | X-ray Structure of a Methanococcus jannaschii DNA-Binding Protein: Implications for Antibiotic Resistance in Staphylococcus aureus | | Descriptor: | hypothetical protein MJ223 | | Authors: | Ray, S.S, Bonanno, J.B, Chen, H, de Lencastre, H, Wu, S, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-01-21 | | Release date: | 2002-12-25 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of an M. jannaschii DNA-binding protein: implications for antibiotic resistance in S.
aureus
Proteins, 50, 2002
|
|
1KVZ
 
 | | Solution Structure of Cytotoxic RC-RNase4 | | Descriptor: | RC-RNase4 | | Authors: | Hsu, C.-H, Liao, Y.-D, Chen, L.-W, Wu, S.-H, Chen, C. | | Deposit date: | 2002-01-28 | | Release date: | 2002-07-28 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Cytotoxic RNase 4 from the Oocytes of Bullfrog Rana Catesbeiana
J.MOL.BIOL., 326, 2003
|
|
1M58
 
 | | Solution Structure of Cytotoxic RC-RNase2 | | Descriptor: | RC-RNase2 ribonuclease | | Authors: | Hsu, C.-H, Liao, Y.-D, Wu, S.-H, Chen, C. | | Deposit date: | 2002-07-09 | | Release date: | 2003-01-09 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C resonance assignments and secondary structure determination of the RC-RNase 2 from oocytes of bullfrog Rana catesbeiana.
J.Biomol.Nmr, 19, 2001
|
|
6KLA
 
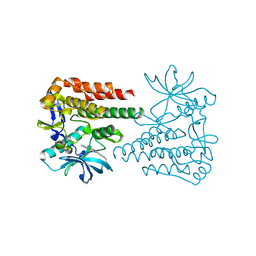 | | Crystal structure of human c-KIT kinase domain in complex with compound 15a | | Descriptor: | Mast/stem cell growth factor receptor Kit, N-[6-(4-ethylpiperazin-1-yl)-2-methyl-pyrimidin-4-yl]-5-pyridin-4-yl-1,3-thiazol-2-amine | | Authors: | Wu, T.S, Peng, Y.H, Hsueh, C.C, Wu, S.Y. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Identification of a Multitargeted Tyrosine Kinase Inhibitor for the Treatment of Gastrointestinal Stromal Tumors and Acute Myeloid Leukemia.
J.Med.Chem., 62, 2019
|
|
1JLZ
 
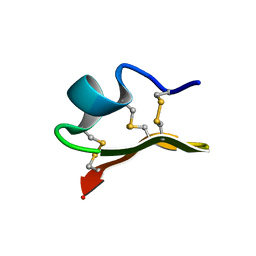 | | Solution Structure of a K+-Channel Blocker from the Scorpion Toxin of Tityus cambridgei | | Descriptor: | Tityustoxin alpha-KTx | | Authors: | Wang, I, Wu, S.-H, Chang, H.-K, Shieh, R.-C, Yu, H.-M, Chen, C. | | Deposit date: | 2001-07-17 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a K(+)-channel blocker from the scorpion Tityus cambridgei.
Protein Sci., 11, 2002
|
|
3JAF
 
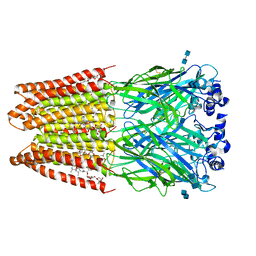 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, glycine/ivermectin-bound state | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.801 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
8IQ9
 
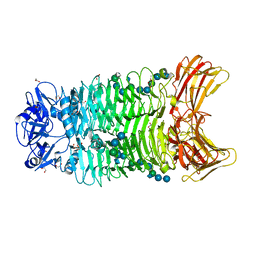 | | Crystal structure of trimeric K2-2 TSP in complex with tetrasaccharide and octasaccharide | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, K2-2 TSP, ... | | Authors: | Ye, T.J, Ko, T.P, Huang, K.F, Wu, S.H. | | Deposit date: | 2023-03-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Klebsiella pneumoniae K2 capsular polysaccharide degradation by a bacteriophage depolymerase does not require trimer formation.
Mbio, 15, 2024
|
|
8IQE
 
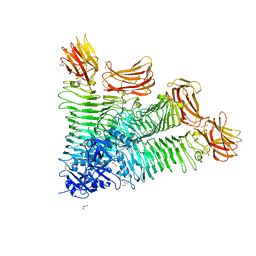 | | Crystal structure of tetrameric K2-2 TSP | | Descriptor: | GLYCEROL, K2-VCL6 TSP | | Authors: | Ye, T.J, Huang, K.F, Tu, I.F, Lee, I.M, Chang, Y.P, Wu, S.H. | | Deposit date: | 2023-03-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Klebsiella pneumoniae K2 capsular polysaccharide degradation by a bacteriophage depolymerase does not require trimer formation.
Mbio, 15, 2024
|
|
3TAT
 
 | | TYROSINE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, TYROSINE AMINOTRANSFERASE | | Authors: | Ko, T.P, Yang, W.Z, Wu, S.P, Tsai, H, Yuan, H.S. | | Deposit date: | 1998-08-12 | | Release date: | 1999-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and preliminary crystallographic analysis of the Escherichia coli tyrosine aminotransferase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
8I6V
 
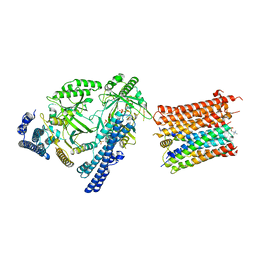 | | Cryo-EM structure of the polyphosphate polymerase VTC complex(Vtc4/Vtc3/Vtc1) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mayer, A, Wu, S, Ye, S. | | Deposit date: | 2023-01-29 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structure of the polyphosphate polymerase VTC reveals coupling of polymer synthesis to membrane transit.
Embo J., 42, 2023
|
|
