1MIS
 
 | |
3D45
 
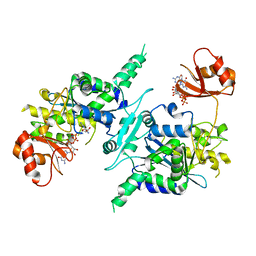 | | Crystal structure of mouse PARN in complex with m7GpppG | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Poly(A)-specific ribonuclease PARN | | Authors: | Wu, M, Song, H. | | Deposit date: | 2008-05-13 | | Release date: | 2009-03-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of m(7)GpppG binding to poly(A)-specific ribonuclease.
Structure, 17, 2009
|
|
5WTA
 
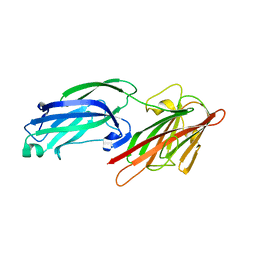 | | Crystal Structure of Staphylococcus aureus SdrE apo form | | Descriptor: | Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
3P13
 
 | |
3N2Y
 
 | |
3P12
 
 | | Crystal Structure of D-ribose Pyranase Sa240 | | Descriptor: | D-ribose pyranase, GLYCEROL | | Authors: | Wu, M, Wang, L, Zang, J. | | Deposit date: | 2010-09-29 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Sa240: A ribose pyranase homolog with partial active site from Staphylococcus aureus
J.Struct.Biol., 174, 2011
|
|
4B5Q
 
 | | The lytic polysaccharide monooxygenase GH61D structure from the basidiomycota fungus Phanerochaete chrysosporium | | Descriptor: | COPPER (II) ION, GLYCEROL, GLYCOSIDE HYDROLASE FAMILY 61 PROTEIN D, ... | | Authors: | Wu, M, Beckham, G.T, Larsson, A.M, Ishida, T, Kim, S, Crowley, M.F, Payne, C.M, Horn, S.J, Westereng, B, Stahlberg, J, Eijsink, V.G.H, Sandgren, M. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Computational Characterization of the Lytic Polysaccharide Monooxygenase Gh61D from the Basidiomycota Fungus Phanerochaete Chrysosporium
J.Biol.Chem., 288, 2013
|
|
4AX6
 
 | | HYPOCREA JECORINA CEL6A D221A MUTANT SOAKED WITH 6-CHLORO-4- PHENYLUMBELLIFERYL-BETA-CELLOBIOSIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloranyl-7-oxidanyl-4-phenyl-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-06-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
4AVN
 
 | | Thermobifida fusca cellobiohydrolase Cel6B catalytic mutant D226A- S232A cocrystallized with cellobiose | | Descriptor: | CALCIUM ION, CELLOBIOHYDROLASE. GLYCOSYL HYDROLASE FAMILY 6, beta-D-glucopyranose, ... | | Authors: | Wu, M, Vuong, T.V, Wilson, D.B, Sandgren, M, Stahlberg, J, Hansson, H. | | Deposit date: | 2012-05-28 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Loop Motions Important to Product Expulsion in the Thermobifida Fusca Glycoside Hydrolase Family 6 Cellobiohydrolase from Structural and Computational Studies.
J.Biol.Chem., 288, 2013
|
|
4AX7
 
 | | Hypocrea jecorina Cel6A D221A mutant soaked with 4-Methylumbelliferyl- beta-D-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-hydroxy-4-methyl-2H-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-06-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
4AVO
 
 | | Thermobifida fusca cellobiohydrolase Cel6B catalytic mutant D274A cocrystallized with cellobiose | | Descriptor: | ACETATE ION, BETA-1,4-EXOCELLULASE, CALCIUM ION, ... | | Authors: | Wu, M, Vuong, T.V, Wilson, D.B, Sandgren, M, Stahlberg, J, Hansson, H. | | Deposit date: | 2012-05-28 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Loop Motions Important to Product Expulsion in the Thermobifida Fusca Glycoside Hydrolase Family 6 Cellobiohydrolase from Structural and Computational Studies.
J.Biol.Chem., 288, 2013
|
|
4AU0
 
 | | Hypocrea jecorina Cel6A D221A mutant soaked with 6-chloro-4- methylumbelliferyl-beta-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-7-hydroxy-4-methyl-2H-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
7T3F
 
 | | Development of BRD4 inhibitors as arsenicals antidotes | | Descriptor: | 4-fluoro-3-methyl-N-(3-methyl-2-oxo-1,2,3,4-tetrahydroquinazolin-6-yl)benzene-1-sulfonamide, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Wu, M, Yatchang, M, Mathew, B, Zhai, L, Ruiz, P, Bostwick, R, Augelli-Szafran, C.E, Suto, M.J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Development of BRD4 inhibitors as anti-inflammatory agents and antidotes for arsenicals.
Bioorg.Med.Chem.Lett., 64, 2022
|
|
5WTB
 
 | | Complex Structure of Staphylococcus aureus SdrE with human complement factor H | | Descriptor: | Peptide from Complement factor H, Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
7T91
 
 | |
5YCO
 
 | | Complex structure of PCNA with UHRF2 | | Descriptor: | E3 ubiquitin-protein ligase UHRF2, GLYCEROL, Proliferating cell nuclear antigen, ... | | Authors: | Wu, M, Chen, W, Hang, T, Wang, C, Zhang, X, Zang, J. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structure insights into the molecular mechanism of the interaction between UHRF2 and PCNA.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
2A1R
 
 | | Crystal structure of PARN nuclease domain | | Descriptor: | 5'-R(*AP*AP*A)-3', Poly(A)-specific ribonuclease PARN | | Authors: | Wu, M, Song, H. | | Deposit date: | 2005-06-21 | | Release date: | 2005-12-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into poly(A) binding and catalytic mechanism of human PARN
Embo J., 24, 2005
|
|
2A1S
 
 | | Crystal structure of native PARN nuclease domain | | Descriptor: | Poly(A)-specific ribonuclease PARN | | Authors: | Wu, M, Song, H. | | Deposit date: | 2005-06-21 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into poly(A) binding and catalytic mechanism of human PARN
Embo J., 24, 2005
|
|
6W0T
 
 | |
6VUQ
 
 | | Crystal structure of CHIKV nsP3 macrodomain soaked with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Nonstructural polyprotein | | Authors: | Wu, M. | | Deposit date: | 2020-02-16 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design.
Plos One, 16, 2021
|
|
6V20
 
 | |
6V21
 
 | |
1YHN
 
 | | Structure basis of RILP recruitment by Rab7 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rab interacting lysosomal protein, ... | | Authors: | Wu, M, Wang, T, Hong, W, Song, H. | | Deposit date: | 2005-01-10 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for recruitment of RILP by small GTPase Rab7.
Embo J., 24, 2005
|
|
1R4A
 
 | | Crystal Structure of GTP-bound ADP-ribosylation Factor Like Protein 1 (Arl1) and GRIP Domain of Golgin245 COMPLEX | | Descriptor: | ADP-ribosylation factor-like protein 1, Golgi autoantigen, golgin subfamily A member 4, ... | | Authors: | Wu, M, Lu, L, Hong, W, Song, H. | | Deposit date: | 2003-10-04 | | Release date: | 2004-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4KJS
 
 | | Structure of native YfkE | | Descriptor: | cation exchanger YfkE | | Authors: | Wu, M, Tong, S, Zheng, L. | | Deposit date: | 2013-05-03 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of Ca2+/H+ antiporter protein YfkE reveals the mechanisms of Ca2+ efflux and its pH regulation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
