6RPC
 
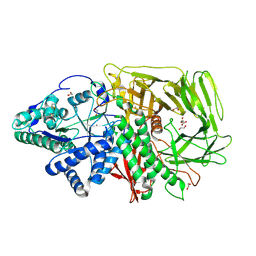 | | GOLGI ALPHA-MANNOSIDASE II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-mannosidase 2, SUCCINIC ACID, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, Debets, M, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6RS0
 
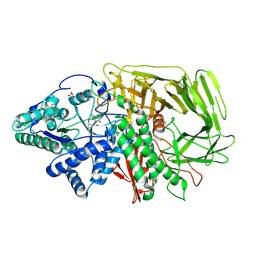 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3S,4R,5R)-1-(2-(Benzyloxy)ethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol | | Descriptor: | (2~{S},3~{S},4~{R})-2-(hydroxymethyl)-1-(2-phenylmethoxyethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
7PSK
 
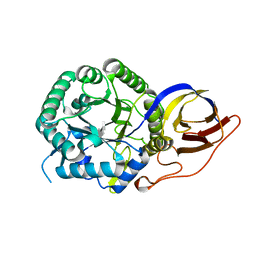 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor GR109 | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Beta-glucuronidase | | Authors: | Armstrong, Z, Wu, L, Davies, G.J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PSI
 
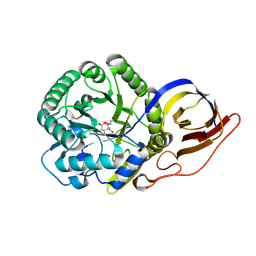 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor ME727 | | Descriptor: | (2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Beta-glucuronidase, SULFATE ION | | Authors: | Armstrong, Z, Wu, L, Davies, G.J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PSH
 
 | |
7PSJ
 
 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor VL166 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Beta-glucuronidase | | Authors: | Armstrong, Z, Wu, L, Davies, G.J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PLS
 
 | | Cryo-EM structures of human fucosidase FucA1 reveal insight into substate recognition and catalysis. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tissue alpha-L-fucosidase | | Authors: | Armstrong, Z, Meek, R.W, Wu, L, Blaza, J.N, Davies, G.J. | | Deposit date: | 2021-09-01 | | Release date: | 2022-08-10 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structures of human fucosidase FucA1 reveal insight into substrate recognition and catalysis.
Structure, 30, 2022
|
|
7PM4
 
 | | Cryo-EM structures of human fucosidase FucA1 reveal insight into substate recognition and catalysis. | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Tissue alpha-L-fucosidase | | Authors: | Armstrong, Z, Meek, R.W, Wu, L, Blaza, J.N, Davies, G.J. | | Deposit date: | 2021-09-01 | | Release date: | 2022-08-10 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structures of human fucosidase FucA1 reveal insight into substrate recognition and catalysis.
Structure, 30, 2022
|
|
1PA9
 
 | | Yersinia Protein-Tyrosine Phosphatase complexed with pNCS (Yop51,Pasteurella X,Ptpase,Yop51delta162) (Catalytic Domain, Residues 163-468) Mutant With Cys 235 Replaced By Arg (C235r) | | Descriptor: | N,4-DIHYDROXY-N-OXO-3-(SULFOOXY)BENZENAMINIUM, Protein-tyrosine phosphatase yopH | | Authors: | Sun, J.P, Wu, L, Fedorov, A.A, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2003-05-13 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Yersinia protein-tyrosine phosphatase YopH complexed
with a specific small molecule inhibitor
J.BIOL.CHEM., 278, 2003
|
|
4DGX
 
 | | LEOPARD Syndrome-Associated SHP2/Y279C mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Xu, J, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L.N, Liu, S.J, Zhang, Z.Y. | | Deposit date: | 2012-01-27 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights into LEOPARD Syndrome-Associated SHP2 Mutations.
J.Biol.Chem., 288, 2013
|
|
4DGP
 
 | | The wild-type Src homology 2 (SH2)-domain containing protein tyrosine phosphatase-2 (SHP2) | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Xu, J, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L.N, Liu, S.J, Zhang, Z.Y. | | Deposit date: | 2012-01-26 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights into LEOPARD Syndrome-Associated SHP2 Mutations.
J.Biol.Chem., 288, 2013
|
|
8CE3
 
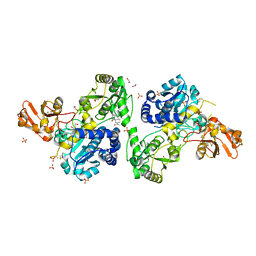 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with 3D fragment 2548 | | Descriptor: | (3~{S})-1-ethanoyl-3-(4-methylphenyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Gilio, A.K, Darby, J.F, Wu, L, Davies, G.J. | | Deposit date: | 2023-02-01 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Modular 3-D Fragment Collection: Design, Synthesis and Screening
To Be Published
|
|
7E1Z
 
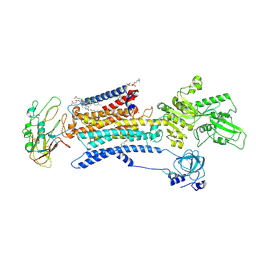 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7E20
 
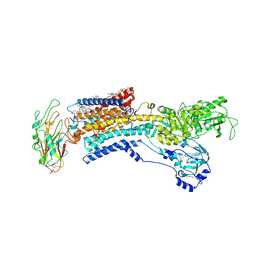 | | Cryo EM structure of a K+-bound Na+,K+-ATPase in the E2 state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7E21
 
 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state with ATP-gamma-S | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
3CKX
 
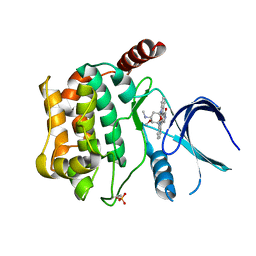 | | Crystal structure of sterile 20-like kinase 3 (MST3, STK24) in complex with staurosporine | | Descriptor: | STAUROSPORINE, Serine/threonine-protein kinase 24 | | Authors: | Antonysamy, S.S, Burley, S.K, Buchanan, S, Chau, F, Feil, I, Wu, L, Sauder, J.M, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-17 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of sterile 20-like kinase 3 (MST3, STK24) in complex with staurosporine.
To be Published
|
|
3O5X
 
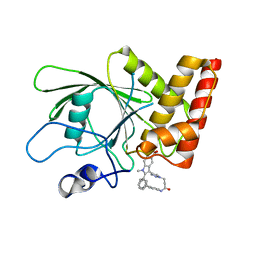 | | Crystal structure of the oncogenic tyrosine phosphatase SHP2 complexed with a salicylic acid-based small molecule inhibitor | | Descriptor: | 3-{1-[3-(biphenyl-4-ylamino)-3-oxopropyl]-1H-1,2,3-triazol-4-yl}-6-hydroxy-1-methyl-2-phenyl-1H-indole-5-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.-Y, Zhang, X, He, Y, Liu, S, Yu, Z, Jiang, Z, Yang, Z, Dong, Y, Nabinger, S.C, Wu, L, Gunawan, A.M, Wang, L, Chan, R.J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salicylic acid based small molecule inhibitor for the oncogenic Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2).
J.Med.Chem., 53, 2010
|
|
7V2T
 
 | | The complex structure of SoBcmB and its natural precursor 2 | | Descriptor: | (3S,6S)-3-((R)-2,3-dihydroxy-2-methylpropyl)-6-((S)-4-hydroxybutan-2-yl)piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Zhou, J.H, Wu, L. | | Deposit date: | 2021-08-09 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20005679 Å) | | Cite: | The complex structure of SoBcmB and its its natural precursor 2
To Be Published
|
|
5L77
 
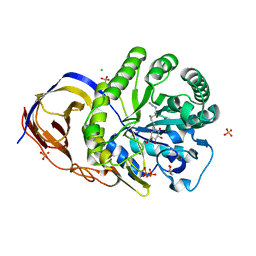 | | A glycoside hydrolase mutant with an unreacted activity based probe bound | | Descriptor: | (1~{R},2~{S},3~{R},4~{S},5~{S},6~{R})-7-[8-[(azanylidene-{4}-azanylidene)amino]octyl]-3,4,5-tris(oxidanyl)-7-azabicyclo[4.1.0]heptane-2-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jin, Y, Wu, L, Jiang, J.B, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2016-06-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
3TJU
 
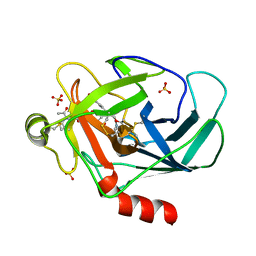 | | Crystal structure of human granzyme H with an inhibitor | | Descriptor: | Ac-PTSY-CMK inhibitor, Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
3TK9
 
 | | Crystal structure of human granzyme H | | Descriptor: | Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
3TJV
 
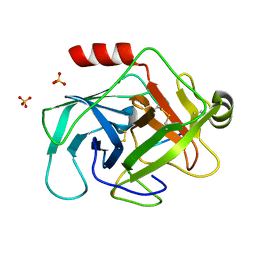 | | Crystal structure of human granzyme H with a peptidyl substrate | | Descriptor: | Granzyme H, PTSYAGDDSG, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
8GTP
 
 | | cryo-EM structure of Omicron BA.5 S protein in complex with XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
8GTO
 
 | | cryo-EM structure of Omicron BA.5 S protein in complex with XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
8GTQ
 
 | | cryo-EM structure of Omicron BA.5 S protein in complex with S2L20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
