4YNZ
 
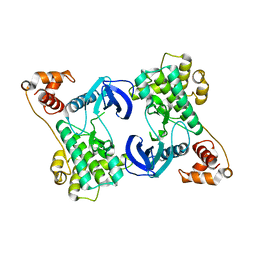 | | Structure of the N-terminal domain of SAD | | Descriptor: | Serine/threonine-protein kinase BRSK2 | | Authors: | Wu, J.X, Wang, J, Chen, L, Wang, Z.X, Wu, J.W. | | Deposit date: | 2015-03-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the mechanism of synergistic autoinhibition of SAD kinases
Nat Commun, 6, 2015
|
|
4YOM
 
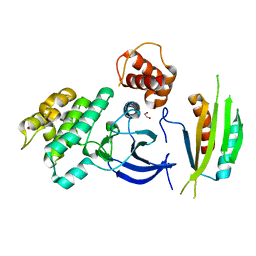 | | Structure of SAD kinase | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase BRSK2 | | Authors: | Wu, J.X, Wang, J, Chen, L, Wang, Z.X, Wu, J.W. | | Deposit date: | 2015-03-12 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insight into the mechanism of synergistic autoinhibition of SAD kinases
Nat Commun, 6, 2015
|
|
8XB4
 
 | |
8XB2
 
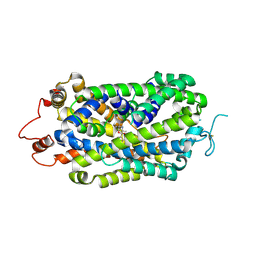 | | Structure of radafaxine-bound state of the human Norepinephrine Transporter | | Descriptor: | (2~{S},3~{S})-2-(3-chlorophenyl)-3,5,5-trimethyl-morpholin-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GFP-MBP-solute carrier family 6 member 2,Maltose/maltodextrin-binding periplasmic protein,Sodium-dependent noradrenaline transporter,Maltose/maltodextrin-binding periplasmic protein,Sodium-dependent noradrenaline transporter | | Authors: | Wu, J.X, Ji, W.M. | | Deposit date: | 2023-12-05 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Substrate binding and inhibition mechanism of norepinephrine transporter
To Be Published
|
|
5IRI
 
 | |
7D3F
 
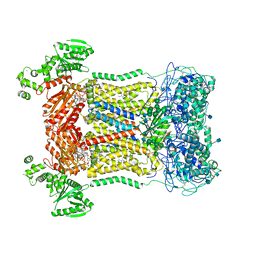 | | Cryo-EM structure of human DUOX1-DUOXA1 in high-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dual oxidase 1, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
8XB3
 
 | |
7D3E
 
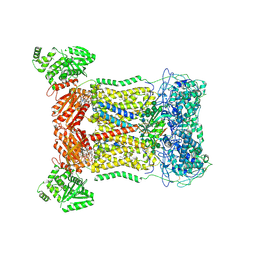 | | Cryo-EM structure of human DUOX1-DUOXA1 in low-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dual oxidase 1, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
5YKF
 
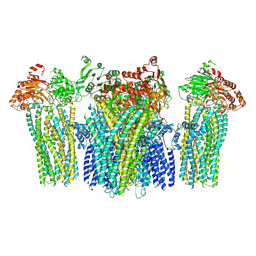 | |
5YKG
 
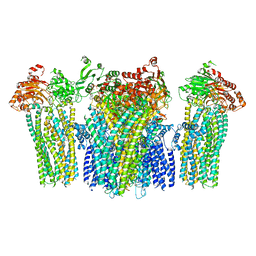 | |
5YKE
 
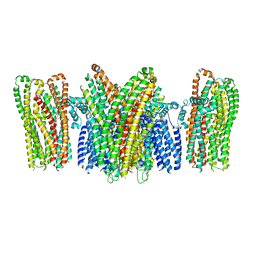 | |
5YW9
 
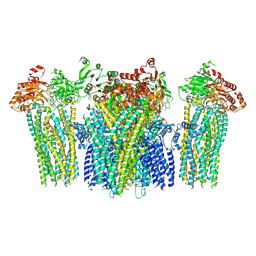 | |
5YWB
 
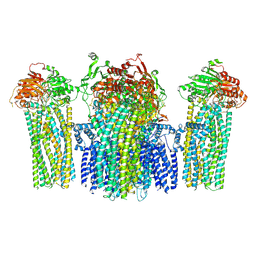 | |
5YWC
 
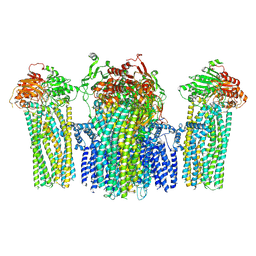 | |
5YW8
 
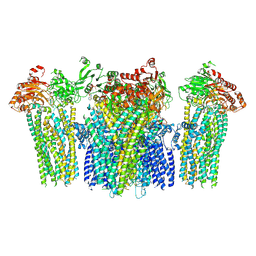 | |
5Z1F
 
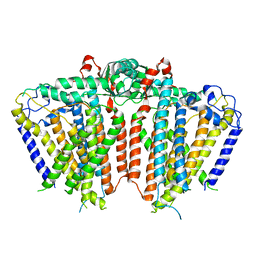 | |
5YWD
 
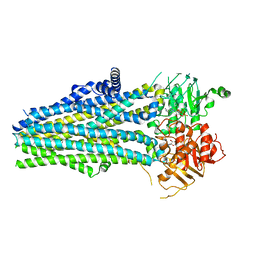 | |
5YWA
 
 | |
5YW7
 
 | |
6JPF
 
 | |
