2XWA
 
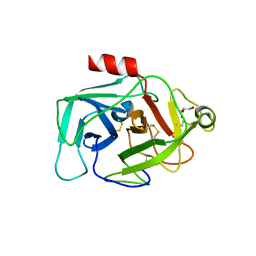 | | Crystal Structure of Complement Factor D Mutant R202A | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
1SGL
 
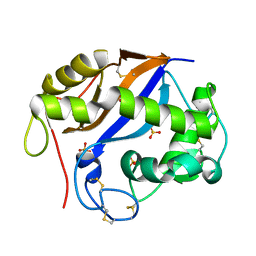 | | The three-dimensional structure and X-ray sequence reveal that trichomaglin is a novel S-like ribonuclease | | Descriptor: | SULFATE ION, trichomaglin | | Authors: | Gan, J.-H, Yu, L, Wu, J, Xu, H, Choudhary, J.S, Blackstock, W.P, Liu, W.-Y, Xia, Z.-X. | | Deposit date: | 2004-02-24 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structure and X-ray sequence reveal that trichomaglin is a novel S-like ribonuclease.
Structure, 12, 2004
|
|
7V2W
 
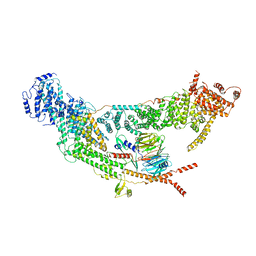 | | protomer structure from the dimer of yeast THO complex | | Descriptor: | Protein TEX1, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
3RES
 
 | |
2XGW
 
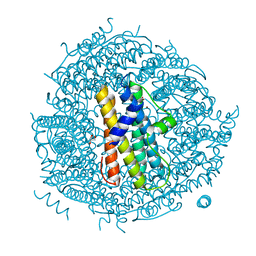 | | ZINC-BOUND CRYSTAL STRUCTURE OF STREPTOCOCCUS PYOGENES DPR | | Descriptor: | CHLORIDE ION, GLYCEROL, PEROXIDE RESISTANCE PROTEIN, ... | | Authors: | Haikarainen, T, Tsou, C.-C, Wu, J.-J, Papageorgiou, A.C. | | Deposit date: | 2010-06-08 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization and Biological Implications of Di-Zinc Binding in the Ferroxidase Center of Streptococcus Pyogenes Dpr.
Biochem.Biophys.Res.Commun., 398, 2010
|
|
5GJQ
 
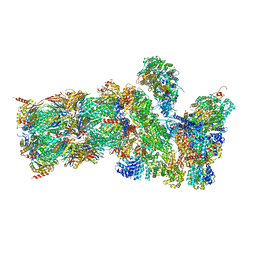 | | Structure of the human 26S proteasome bound to USP14-UbAl | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Huang, X.L, Luan, B, Wu, J.P, Shi, Y.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-08-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | An atomic structure of the human 26S proteasome.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5GJR
 
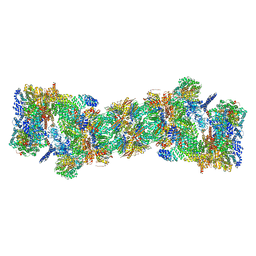 | | An atomic structure of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Huang, X.L, Luan, B, Wu, J.P, Shi, Y.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An atomic structure of the human 26S proteasome.
Nat. Struct. Mol. Biol., 23, 2016
|
|
6L7O
 
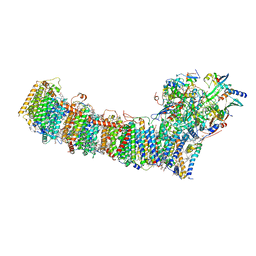 | | cryo-EM structure of cyanobacteria Fd-NDH-1L complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Zhang, C, Shuai, J, Wu, J, Lei, M. | | Deposit date: | 2019-11-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into NDH-1 mediated cyclic electron transfer.
Nat Commun, 11, 2020
|
|
6LBS
 
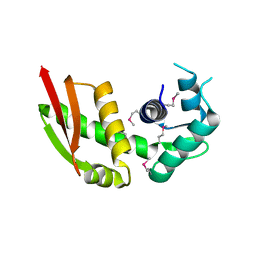 | | Crystal structure of yeast Stn1 | | Descriptor: | KLLA0C11825p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3NQX
 
 | | Crystal structure of vibriolysin MCP-02 mature enzyme, a zinc metalloprotease from M4 family | | Descriptor: | CALCIUM ION, Secreted metalloprotease Mcp02, ZINC ION | | Authors: | Gao, X, Wang, J, Wu, J.-W, Zhang, Y.-Z. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the autoprocessing of zinc metalloproteases in the thermolysin family
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NQZ
 
 | | Crystal structure of the autoprocessed Vibriolysin MCP-02 with E369A mutation | | Descriptor: | CALCIUM ION, Secreted metalloprotease Mcp02, ZINC ION | | Authors: | Gao, X, Wang, J, Chen, L, Wu, J.-W, Zhang, Y.-Z. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for the autoprocessing of zinc metalloproteases in the thermolysin family
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1TP7
 
 | | Crystal Structure of the RNA-dependent RNA Polymerase from Human Rhinovirus 16 | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, Genome polyprotein, SULFATE ION | | Authors: | Appleby, T.C, Luecke, H, Shim, J.H, Wu, J.Z, Cheney, I.W, Zhong, W, Vogeley, L, Hong, Z, Yao, N. | | Deposit date: | 2004-06-15 | | Release date: | 2005-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of complete rhinovirus RNA polymerase suggests front loading of protein primer.
J.Virol., 79, 2005
|
|
1LCY
 
 | | Crystal Structure of the Mitochondrial Serine Protease HtrA2 | | Descriptor: | HtrA2 serine protease | | Authors: | Li, W, Srinivasula, S.M, Chai, J, Li, P, Wu, J.W, Zhang, Z, Alnemri, E.S, Shi, Y. | | Deposit date: | 2002-04-07 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi.
Nat.Struct.Biol., 9, 2002
|
|
3NQY
 
 | | Crystal structure of the autoprocessed complex of Vibriolysin MCP-02 with a single point mutation E346A | | Descriptor: | CALCIUM ION, Secreted metalloprotease Mcp02, ZINC ION | | Authors: | Gao, X, Wang, J, Wu, J.-W, Zhang, Y.-Z. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the autoprocessing of zinc metalloproteases in the thermolysin family
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1NB8
 
 | | Structure of the catalytic domain of USP7 (HAUSP) | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Li, P, Li, M, Li, W, Yao, T, Wu, J.-W, Gu, W, Cohen, R.E, Shi, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde
Cell(Cambridge,Mass.), 111, 2002
|
|
1SJ0
 
 | | Human Estrogen Receptor Alpha Ligand-binding Domain in Complex with the Antagonist Ligand 4-D | | Descriptor: | (2S,3R)-2-(4-(2-(PIPERIDIN-1-YL)ETHOXY)PHENYL)-2,3-DIHYDRO-3-(4-HYDROXYPHENYL)BENZO[B][1,4]OXATHIIN-6-OL, Estrogen receptor | | Authors: | Kim, S, Wu, J.Y, Birzin, E.T, Chan, W, Pai, L.Y, Yang, Y.T, Mosley, R.T, Fitzgerald, P.M, Sharma, N, DiNinno, F, Rohrer, S.P, Schaeffer, J.M, Hammond, M.L. | | Deposit date: | 2004-03-02 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Estrogen Receptor Ligands. II. Discovery of Benzoxathiins as Potent, Selective Estrogen Receptor alpha Modulators.
J.Med.Chem., 47, 2004
|
|
1LQX
 
 | | Crystal structure of V45E mutant of cytochrome b5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Gan, J.-H, Wu, J, Wang, Z.-Q, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2002-05-14 | | Release date: | 2002-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of V45E and V45Y mutants and structure comparison of a variety of cytochrome b5 mutants.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1LR6
 
 | | Crystal structure of V45Y mutant of cytochrome b5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Gan, J.-H, Wu, J, Wang, Z.-Q, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2002-05-15 | | Release date: | 2002-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of V45E and V45Y mutants and structure comparison of a variety of cytochrome b5 mutants.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3G0E
 
 | | KIT kinase domain in complex with sunitinib | | Descriptor: | Mast/stem cell growth factor receptor, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Gajiwala, K.S, Wu, J.C, Lunney, E.A, Gemetri, G.D. | | Deposit date: | 2009-01-27 | | Release date: | 2009-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3BCH
 
 | |
5BW2
 
 | | X-ray crystal structure of Aplysia californica acetylcholine binding protein (Ac-AChBP) Y55W in complex with 2-Pyridin-3-yl-1-aza-bicyclo[2.2.2]octane; 2-(3-pyridyl)quinuclidine; 2-PQ (TI-4699) | | Descriptor: | (2R)-2-(pyridin-3-yl)-1-azabicyclo[2.2.2]octane, Soluble acetylcholine receptor | | Authors: | Bobango, J, Sankaran, B, Park, J.F, Wu, J, Talley, T.T. | | Deposit date: | 2015-06-05 | | Release date: | 2015-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Comparisons of Binding Affinities for Neuronal Nicotinic Receptors (NNRs) and AChBPs
To Be Published
|
|
1CXR
 
 | |
4YE6
 
 | |
4ZK1
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein (LsAChBP) in Complex with 3-Pyrrolylmethylene Anabaseine | | Descriptor: | (3E)-3-(1H-pyrrol-3-ylmethylidene)-3,4,5,6-tetrahydro-2,3'-bipyridine, Acetylcholine-binding protein, PHOSPHATE ION | | Authors: | Bobango, J, Wu, J, Talley, T.T. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein (LsAChBP) in Complex with 3-Pyrrolylmethylene Anabaseine
To be Published
|
|
6JPB
 
 | | Rabbit Cav1.1-Diltiazem Complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
